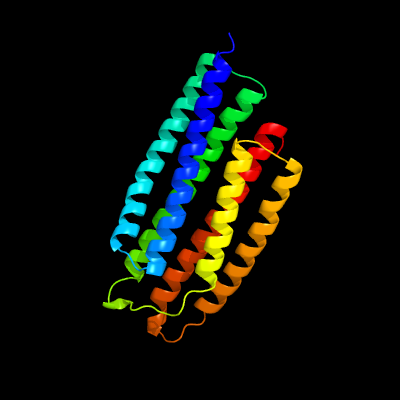
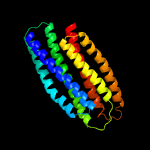
1 d1sumb_
100.0
17
Fold: Spectrin repeat-likeSuperfamily: PhoU-likeFamily: PhoU-like
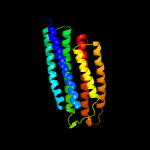
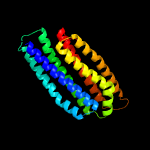
2 d1t72a_
100.0
24
Fold: Spectrin repeat-likeSuperfamily: PhoU-likeFamily: PhoU-like
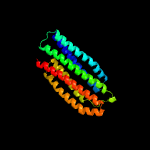
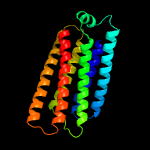
3 d1xwma_
100.0
29
Fold: Spectrin repeat-likeSuperfamily: PhoU-likeFamily: PhoU-like
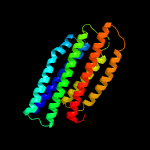
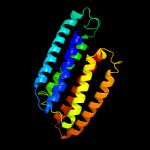
4 c4q25B_
100.0
23
PDB header: unknown functionChain: B: PDB Molecule: phosphate-specific transport system accessory protein phouPDBTitle: crystal structure of phou from pseudomonas aeruginosa
5 c2i0mA_
100.0
23
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: phosphate transport system protein phou;PDBTitle: crystal structure of the phosphate transport system regulatory protein2 phou from streptococcus pneumoniae
6 c3l39A_
99.8
9
PDB header: phosphate-binding proteinChain: A: PDB Molecule: putative phou-like phosphate regulatory protein;PDBTitle: crystal structure of putative phou-like phosphate regulatory protein2 (bt4638) from bacteroides thetaiotaomicron vpi-5482 at 1.93 a3 resolution
7 c2oltB_
99.8
15
PDB header: unknown functionChain: B: PDB Molecule: hypothetical protein;PDBTitle: crystal structure of a phou-like protein (so_3770) from shewanella2 oneidensis mr-1 at 2.00 a resolution
8 d1vcta1
99.6
22
Fold: Spectrin repeat-likeSuperfamily: PhoU-likeFamily: PhoU-like
9 c2bknA_
99.3
22
PDB header: membrane proteinChain: A: PDB Molecule: hypothetical protein ph0236;PDBTitle: structure analysis of unknown function protein
10 c5vwvB_
38.1
25
PDB header: apoptosisChain: B: PDB Molecule: bcl-2-like protein 11;PDBTitle: bak core latch dimer in complex with bim-bh3 - cubic
11 c5vwwC_
37.6
25
PDB header: apoptosisChain: C: PDB Molecule: bcl-2-like protein 11;PDBTitle: bak core latch dimer in complex with bim-rt - tetragonal
12 c5vwwD_
34.5
25
PDB header: apoptosisChain: D: PDB Molecule: bcl-2-like protein 11;PDBTitle: bak core latch dimer in complex with bim-rt - tetragonal
13 c1pq1B_
29.3
25
PDB header: apoptosisChain: B: PDB Molecule: bcl2-like protein 11;PDBTitle: crystal structure of bcl-xl/bim
14 c3izcw_
28.2
16
PDB header: ribosomeChain: W: PDB Molecule: 60s ribosomal protein rpl22 (l22e);PDBTitle: localization of the large subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
15 c5ua4B_
27.4
25
PDB header: apoptosisChain: B: PDB Molecule: bh3-interacting domain death agonist;PDBTitle: crystal structure of a179l:bid bh3 complex
16 c5vmoB_
22.5
17
PDB header: viral protein/apoptosisChain: B: PDB Molecule: bcl-2 interacting mediator of cell death;PDBTitle: crystal structure of grouper iridovirus giv66:bim complex
17 d1l0oc_
20.9
21
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: Sigma4 domain
18 c1l0oC_
20.9
21
PDB header: protein bindingChain: C: PDB Molecule: sigma factor;PDBTitle: crystal structure of the bacillus stearothermophilus anti-2 sigma factor spoiiab with the sporulation sigma factor3 sigmaf
19 c2k7wB_
20.3
30
PDB header: apoptosisChain: B: PDB Molecule: bcl-2-like protein 11;PDBTitle: bax activation is initiated at a novel interaction site
20 c4qvfB_
18.6
25
PDB header: apoptosisChain: B: PDB Molecule: peptide from bcl-2-like protein 11;PDBTitle: crystal structure of bcl-xl in complex with bim bh3 domain
21 c5wosB_
not modelled
18.6
25
PDB header: viral proteinChain: B: PDB Molecule: bcl-2-like protein 11;PDBTitle: structural and functional insights into canarypox virus cnp0582 regulation of apoptosis
22 c4zieC_
not modelled
18.1
25
PDB header: apoptosisChain: C: PDB Molecule: bcl-2-like protein 11;PDBTitle: crystal structure of core/latch dimer of bax in complex with bimbh3
23 c4d2mB_
not modelled
18.1
25
PDB header: apoptosisChain: B: PDB Molecule: bcl-2-like protein 11;PDBTitle: vaccinia virus f1l bound to bim bh3
24 c4d2mD_
not modelled
18.0
25
PDB header: apoptosisChain: D: PDB Molecule: bcl-2-like protein 11;PDBTitle: vaccinia virus f1l bound to bim bh3
25 c2v6qB_
not modelled
17.9
25
PDB header: apoptosisChain: B: PDB Molecule: bcl-2-like protein 11;PDBTitle: crystal structure of a bhrf-1 : bim bh3 complex
26 c2wh6B_
not modelled
17.8
25
PDB header: apoptosisChain: B: PDB Molecule: bcl-2-like protein 11;PDBTitle: crystal structure of anti-apoptotic bhrf1 in complex with the bim bh32 domain
27 c2pqkB_
not modelled
17.7
25
PDB header: apoptosisChain: B: PDB Molecule: bim bh3 peptide;PDBTitle: x-ray crystal structure of human mcl-1 in complex with bim bh3
28 c2nl9B_
not modelled
17.5
25
PDB header: apoptosisChain: B: PDB Molecule: bcl-2-like protein 11;PDBTitle: crystal structure of the mcl-1:bim bh3 complex
29 c6qfiB_
not modelled
17.5
25
PDB header: apoptosisChain: B: PDB Molecule: bcl-2-like protein 11;PDBTitle: structure of human mcl-1 in complex with bim bh3 peptide
30 c3fdlB_
not modelled
17.2
25
PDB header: apoptosisChain: B: PDB Molecule: bcl-2-like protein 11;PDBTitle: bim bh3 peptide in complex with bcl-xl
31 c3kj2B_
not modelled
17.1
25
PDB header: apoptosisChain: B: PDB Molecule: bcl-2-like protein 11;PDBTitle: mcl-1 in complex with bim bh3 mutant f4ae
32 c3ci9B_
not modelled
16.2
18
PDB header: transcriptionChain: B: PDB Molecule: heat shock factor-binding protein 1;PDBTitle: crystal structure of the human hsbp1
33 d1seta1
not modelled
14.6
17
Fold: Long alpha-hairpinSuperfamily: tRNA-binding armFamily: Seryl-tRNA synthetase (SerRS)
34 c3kz0C_
not modelled
12.6
17
PDB header: apoptosisChain: C: PDB Molecule: mcl-1 specific peptide mb7;PDBTitle: mcl-1 complex with mcl-1-specific selected peptide
35 c3kz0D_
not modelled
12.6
17
PDB header: apoptosisChain: D: PDB Molecule: mcl-1 specific peptide mb7;PDBTitle: mcl-1 complex with mcl-1-specific selected peptide
36 c5l33A_
not modelled
12.1
23
PDB header: de novo proteinChain: A: PDB Molecule: denovo ntf2;PDBTitle: crystal structure of a de novo designed protein with curved beta-sheet
37 d1uptb_
not modelled
11.6
3
Fold: GRIP domainSuperfamily: GRIP domainFamily: GRIP domain
38 c3d7vB_
not modelled
10.8
29
PDB header: apoptosisChain: B: PDB Molecule: bcl-2-like protein 11;PDBTitle: crystal structure of mcl-1 in complex with an mcl-1 selective bh32 ligand
39 d1eqzb_
not modelled
10.2
19
Fold: Histone-foldSuperfamily: Histone-foldFamily: Nucleosome core histones
40 d1kx5d_
not modelled
10.1
21
Fold: Histone-foldSuperfamily: Histone-foldFamily: Nucleosome core histones
41 d1s32d_
not modelled
10.1
21
Fold: Histone-foldSuperfamily: Histone-foldFamily: Nucleosome core histones
42 c3io8D_
not modelled
10.0
30
PDB header: apoptosisChain: D: PDB Molecule: bcl-2-like protein 11;PDBTitle: biml12f in complex with bcl-xl
43 d1nexa1
not modelled
9.7
15
Fold: Skp1 dimerisation domain-likeSuperfamily: Skp1 dimerisation domain-likeFamily: Skp1 dimerisation domain-like
44 c4j7oA_
not modelled
9.6
40
PDB header: cell invasionChain: A: PDB Molecule: putative surface cell antigen sca2;PDBTitle: structure of the n-terminal repeat domain of rickettsia sca2
45 c1nfoA_
not modelled
9.4
11
PDB header: lipid transportChain: A: PDB Molecule: apolipoprotein e2;PDBTitle: apolipoprotein e2 (apoe2, d154a mutation)
46 c3e6sD_
not modelled
9.3
9
PDB header: oxidoreductaseChain: D: PDB Molecule: ferritin;PDBTitle: crystal structure of ferritin soaked with iron from pseudo-nitzschia2 multiseries
47 c3axjB_
not modelled
9.1
9
PDB header: dna binding proteinChain: B: PDB Molecule: translin associated factor x, isoform b;PDBTitle: high resolution crystal structure of c3po
48 d1jmsa1
not modelled
9.1
7
Fold: SAM domain-likeSuperfamily: DNA polymerase beta, N-terminal domain-likeFamily: DNA polymerase beta, N-terminal domain-like
49 c4h63K_
not modelled
8.9
24
PDB header: transcriptionChain: K: PDB Molecule: mediator of rna polymerase ii transcription subunit 11;PDBTitle: structure of the schizosaccharomyces pombe mediator head module
50 c3io8B_
not modelled
8.8
25
PDB header: apoptosisChain: B: PDB Molecule: bcl-2-like protein 11;PDBTitle: biml12f in complex with bcl-xl
51 c6qeqD_
not modelled
8.6
17
PDB header: dna binding proteinChain: D: PDB Molecule: pcff;PDBTitle: pcff from enterococcus faecalis pcf10
52 c3kj0B_
not modelled
8.4
22
PDB header: apoptosisChain: B: PDB Molecule: bcl-2-like protein 11;PDBTitle: mcl-1 in complex with bim bh3 mutant i2dy
53 d1fs1b1
not modelled
8.0
18
Fold: Skp1 dimerisation domain-likeSuperfamily: Skp1 dimerisation domain-likeFamily: Skp1 dimerisation domain-like
54 c3io9B_
not modelled
7.5
25
PDB header: apoptosisChain: B: PDB Molecule: bcl-2-like protein 11;PDBTitle: biml12y in complex with mcl-1
55 c3kgkA_
not modelled
7.4
27
PDB header: chaperoneChain: A: PDB Molecule: arsenical resistance operon trans-acting repressor arsd;PDBTitle: crystal structure of arsd
56 d1fs2b1
not modelled
7.3
18
Fold: Skp1 dimerisation domain-likeSuperfamily: Skp1 dimerisation domain-likeFamily: Skp1 dimerisation domain-like
57 c3kj1B_
not modelled
7.1
17
PDB header: apoptosisChain: B: PDB Molecule: bcl-2-like protein 11;PDBTitle: mcl-1 in complex with bim bh3 mutant i2da
58 d2o02a1
not modelled
7.0
12
Fold: alpha-alpha superhelixSuperfamily: 14-3-3 proteinFamily: 14-3-3 protein
59 d1uptd_
not modelled
7.0
4
Fold: GRIP domainSuperfamily: GRIP domainFamily: GRIP domain
60 c2k6sB_
not modelled
6.9
12
PDB header: protein transportChain: B: PDB Molecule: rab11fip2 protein;PDBTitle: structure of rab11-fip2 c-terminal coiled-coil domain
61 c3p8cF_
not modelled
6.8
11
PDB header: protein bindingChain: F: PDB Molecule: abl interactor 2;PDBTitle: structure and control of the actin regulatory wave complex
62 d2coha1
not modelled
6.7
9
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: CAP C-terminal domain-like
63 c3iz5w_
not modelled
6.7
23
PDB header: ribosomeChain: W: PDB Molecule: 60s ribosomal protein l22 (l22e);PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
64 c5jreC_
not modelled
6.5
9
PDB header: dna binding proteinChain: C: PDB Molecule: neq131;PDBTitle: crystal structure of nec3po in complex with ssdna.
65 c3d1dC_
not modelled
6.4
24
PDB header: nuclear proteinChain: C: PDB Molecule: rna-induced transcriptional silencing complexPDBTitle: hexagonal crystal structure of tas3 c-terminal alpha motif
66 c4dg7D_
not modelled
6.3
13
PDB header: dna binding proteinChain: D: PDB Molecule: gm27569p;PDBTitle: low resolution structure of drosophila translin
67 c3fmtF_
not modelled
6.2
16
PDB header: replication inhibitor/dnaChain: F: PDB Molecule: protein seqa;PDBTitle: crystal structure of seqa bound to dna
68 d1ofcx1
not modelled
6.1
23
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Myb/SANT domain
69 c3ktbD_
not modelled
6.0
27
PDB header: transcription regulatorChain: D: PDB Molecule: arsenical resistance operon trans-acting repressor;PDBTitle: crystal structure of arsenical resistance operon trans-acting2 repressor from bacteroides vulgatus atcc 8482
70 d1t6ua_
not modelled
5.9
8
Fold: Four-helical up-and-down bundleSuperfamily: Nickel-containing superoxide dismutase, NiSODFamily: Nickel-containing superoxide dismutase, NiSOD
71 d1rp3a1
not modelled
5.7
17
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: Sigma3 domain
72 d1q0ga_
not modelled
5.7
8
Fold: Four-helical up-and-down bundleSuperfamily: Nickel-containing superoxide dismutase, NiSODFamily: Nickel-containing superoxide dismutase, NiSOD
73 d1tzyb_
not modelled
5.6
19
Fold: Histone-foldSuperfamily: Histone-foldFamily: Nucleosome core histones
74 d1ku2a1
not modelled
5.6
23
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: Sigma3 domain
75 d2ovra1
not modelled
5.6
19
Fold: Skp1 dimerisation domain-likeSuperfamily: Skp1 dimerisation domain-likeFamily: Skp1 dimerisation domain-like
76 d1x92a_
not modelled
5.4
16
Fold: SIS domainSuperfamily: SIS domainFamily: mono-SIS domain
77 c2xa0C_
not modelled
5.3
25
PDB header: apoptosisChain: C: PDB Molecule: apoptosis regulator bax;PDBTitle: crystal structure of bcl-2 in complex with a bax bh32 peptide
78 c5toiB_
not modelled
5.3
14
PDB header: viral proteinChain: B: PDB Molecule: polymerase cofactor vp35;PDBTitle: crystal structure of the marburg virus vp35 oligomerization domain2 p4222
79 c4d18I_
not modelled
5.3
8
PDB header: signaling proteinChain: I: PDB Molecule: cop9 signalosome complex subunit 1;PDBTitle: crystal structure of the cop9 signalosome
80 c4khaA_
not modelled
5.3
14
PDB header: chaperone/nuclear proteinChain: A: PDB Molecule: spt16m-histone h2b 1.1 chimera;PDBTitle: structural basis of histone h2a-h2b recognition by the essential2 chaperone fact
81 d1fpoa2
not modelled
5.2
19
Fold: Open three-helical up-and-down bundleSuperfamily: HSC20 (HSCB), C-terminal oligomerisation domainFamily: HSC20 (HSCB), C-terminal oligomerisation domain
82 c5xauC_
not modelled
5.2
13
PDB header: cell adhesionChain: C: PDB Molecule: laminin subunit gamma-1;PDBTitle: crystal structure of integrin binding fragment of laminin-511