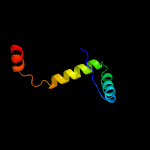
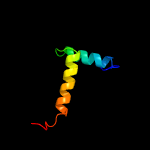
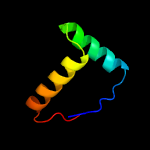
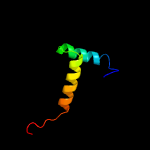
1 c1q5vB_
95.1
27
PDB header: transcriptionChain: B: PDB Molecule: nickel responsive regulator;PDBTitle: apo-nikr
2 c5x3tA_
95.1
24
PDB header: antitoxin/toxinChain: A: PDB Molecule: antitoxin vapb26;PDBTitle: vapbc from mycobacterium tuberculosis
3 c2bj3D_
94.9
14
PDB header: transcriptionChain: D: PDB Molecule: nickel responsive regulator;PDBTitle: nikr-apo
4 c2k5jB_
94.0
17
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein yiif;PDBTitle: solution structure of protein yiif from shigella flexneri2 serotype 5b (strain 8401) . northeast structural genomics3 consortium target sft1
5 d2bsqe1
93.7
17
Fold: Ribbon-helix-helixSuperfamily: Ribbon-helix-helixFamily: Trafficking protein A-like
6 c3kk4B_
93.6
11
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein bp1543;PDBTitle: uncharacterized protein bp1543 from bordetella pertussis tohama i
7 c2h1oH_
93.3
17
PDB header: gene regulation/dna complexChain: H: PDB Molecule: trafficking protein a;PDBTitle: structure of fitab bound to ir36 dna fragment
8 d2bj7a1
92.9
20
Fold: Ribbon-helix-helixSuperfamily: Ribbon-helix-helixFamily: CopG-like
9 c2ca9B_
91.8
16
PDB header: transcriptionChain: B: PDB Molecule: putative nickel-responsive regulator;PDBTitle: apo-nikr from helicobacter pylori in closed trans-2 conformation
10 d2hzaa1
90.1
30
Fold: Ribbon-helix-helixSuperfamily: Ribbon-helix-helixFamily: CopG-like
11 d2hzab1
87.7
30
Fold: Ribbon-helix-helixSuperfamily: Ribbon-helix-helixFamily: CopG-like
12 c6a7vU_
87.2
23
PDB header: toxin/antitoxinChain: U: PDB Molecule: antitoxin vapb11;PDBTitle: crystal structure of mycobacterium tuberculosis vapbc11 toxin-2 antitoxin complex
13 c1xrxD_
73.2
29
PDB header: replication inhibitorChain: D: PDB Molecule: seqa protein;PDBTitle: crystal structure of a dna-binding protein
14 d1xrxa1
73.2
29
Fold: Ribbon-helix-helixSuperfamily: Ribbon-helix-helixFamily: SeqA N-terminal domain-like
15 c3h87D_
69.1
29
PDB header: toxin/antitoxinChain: D: PDB Molecule: putative uncharacterized protein;PDBTitle: rv0301 rv0300 toxin antitoxin complex from mycobacterium tuberculosis
16 c2k29A_
67.4
20
PDB header: transcriptionChain: A: PDB Molecule: antitoxin relb;PDBTitle: structure of the dbd domain of e. coli antitoxin relb
17 c6g1nB_
53.3
26
PDB header: antitoxinChain: B: PDB Molecule: antitoxin hicb;PDBTitle: crystal structure of the burkholderia pseudomallei antitoxin hicb
18 c2an7A_
47.9
21
PDB header: dna binding proteinChain: A: PDB Molecule: protein pard;PDBTitle: solution structure of the bacterial antidote pard
19 d1mnta_
43.0
22
Fold: Ribbon-helix-helixSuperfamily: Ribbon-helix-helixFamily: Arc/Mnt-like phage repressors
20 c3fmtF_
42.9
16
PDB header: replication inhibitor/dnaChain: F: PDB Molecule: protein seqa;PDBTitle: crystal structure of seqa bound to dna
21 d2cpga_
not modelled
40.2
23
Fold: Ribbon-helix-helixSuperfamily: Ribbon-helix-helixFamily: CopG-like
22 c1ea4K_
not modelled
40.2
23
PDB header: gene regulation/dnaChain: K: PDB Molecule: transcriptional repressor copg;PDBTitle: transcriptional repressor copg/22bp dsdna complex
23 c1u9pA_
not modelled
24.9
15
PDB header: unknown functionChain: A: PDB Molecule: parc;PDBTitle: permuted single-chain arc
24 c4fxeB_
not modelled
24.8
20
PDB header: toxin/toxin inhibitorChain: B: PDB Molecule: antitoxin relb;PDBTitle: crystal structure of the intact e. coli relbe toxin-antitoxin complex
25 c4p7dA_
not modelled
23.4
21
PDB header: toxinChain: A: PDB Molecule: antitoxin hicb3;PDBTitle: antitoxin hicb3 crystal structure
26 c2mdvB_
not modelled
21.9
25
PDB header: de novo proteinChain: B: PDB Molecule: designed protein;PDBTitle: nmr structure of beta alpha alpha 38
27 c1t01B_
not modelled
20.9
47
PDB header: cell adhesion, structural proteinChain: B: PDB Molecule: talin 1;PDBTitle: vinculin complexed with the vbs1 helix from talin
28 d1wuda1
not modelled
19.8
17
Fold: SAM domain-likeSuperfamily: HRDC-likeFamily: HRDC domain from helicases
29 c6a6xC_
not modelled
19.0
26
PDB header: toxinChain: C: PDB Molecule: antitoxin maze7;PDBTitle: the crystal structure of the mtb maze-mazf-mt9 complex
30 d1s4ka_
not modelled
19.0
22
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: YdiL-like
31 c1syqB_
not modelled
18.3
47
PDB header: cell adhesionChain: B: PDB Molecule: talin 1;PDBTitle: human vinculin head domain vh1, residues 1-258, in complex with human2 talin's vinculin binding site 1, residues 607-636
32 d1a2za_
not modelled
17.9
15
Fold: Phosphorylase/hydrolase-likeSuperfamily: Pyrrolidone carboxyl peptidase (pyroglutamate aminopeptidase)Family: Pyrrolidone carboxyl peptidase (pyroglutamate aminopeptidase)
33 c3qoqC_
not modelled
16.6
16
PDB header: transcription/dnaChain: C: PDB Molecule: alginate and motility regulator z;PDBTitle: crystal structure of the transcription factor amrz in complex with the2 18 base pair amrz1 binding site
34 c4q2uM_
not modelled
16.1
10
PDB header: toxin/toxin repressorChain: M: PDB Molecule: antitoxin dinj;PDBTitle: crystal structure of the e. coli dinj-yafq toxin-antitoxin complex
35 d1iofa_
not modelled
13.9
19
Fold: Phosphorylase/hydrolase-likeSuperfamily: Pyrrolidone carboxyl peptidase (pyroglutamate aminopeptidase)Family: Pyrrolidone carboxyl peptidase (pyroglutamate aminopeptidase)
36 d1zq1c2
not modelled
13.5
21
Fold: DCoH-likeSuperfamily: GAD domain-likeFamily: GAD domain
37 d1iu8a_
not modelled
13.4
20
Fold: Phosphorylase/hydrolase-likeSuperfamily: Pyrrolidone carboxyl peptidase (pyroglutamate aminopeptidase)Family: Pyrrolidone carboxyl peptidase (pyroglutamate aminopeptidase)
38 c4a1qB_
not modelled
13.1
28
PDB header: viral proteinChain: B: PDB Molecule: orf e73;PDBTitle: solution structure of e73 protein from sulfolobus spindle-2 shaped virus ragged hills, a hyperthermophilic3 crenarchaeal virus from yellowstone national park
39 c4aaiB_
not modelled
13.1
28
PDB header: viral proteinChain: B: PDB Molecule: orf e73;PDBTitle: thermostable protein from hyperthermophilic virus ssv-rh
40 c3lacA_
not modelled
12.5
16
PDB header: hydrolaseChain: A: PDB Molecule: pyrrolidone-carboxylate peptidase;PDBTitle: crystal structure of bacillus anthracis pyrrolidone-carboxylate2 peptidase, pcp
41 c3giuA_
not modelled
12.0
25
PDB header: hydrolaseChain: A: PDB Molecule: pyrrolidone-carboxylate peptidase;PDBTitle: 1.25 angstrom crystal structure of pyrrolidone-carboxylate peptidase2 (pcp) from staphylococcus aureus
42 c2jr6A_
not modelled
11.9
17
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: upf0434 protein nma0874;PDBTitle: solution structure of upf0434 protein nma0874. northeast structural2 genomics target mr32
43 c5htlA_
not modelled
11.8
19
PDB header: biosynthetic proteinChain: A: PDB Molecule: msha biogenesis protein mshe;PDBTitle: structure of mshe with cdg
44 c3t98C_
not modelled
11.5
24
PDB header: protein transportChain: C: PDB Molecule: nuclear pore complex protein nup54;PDBTitle: molecular architecture of the transport channel of the nuclear pore2 complex: nup54/nup58
45 d1d8wa_
not modelled
11.4
24
Fold: TIM beta/alpha-barrelSuperfamily: Xylose isomerase-likeFamily: L-rhamnose isomerase
46 c4gxhC_
not modelled
10.4
23
PDB header: hydrolaseChain: C: PDB Molecule: pyrrolidone-carboxylate peptidase;PDBTitle: crystal structure of a pyrrolidone-carboxylate peptidase 1 (target id2 nysgrc-012831) from xenorhabdus bovienii ss-2004
47 c6ejfQ_
not modelled
10.4
11
PDB header: motor proteinChain: Q: PDB Molecule: type iv pilus assembly protein pilf;PDBTitle: thermus thermophilus pilf atpase (apoprotein form)
48 d1auga_
not modelled
9.8
27
Fold: Phosphorylase/hydrolase-likeSuperfamily: Pyrrolidone carboxyl peptidase (pyroglutamate aminopeptidase)Family: Pyrrolidone carboxyl peptidase (pyroglutamate aminopeptidase)
49 d2d6fc2
not modelled
9.6
23
Fold: DCoH-likeSuperfamily: GAD domain-likeFamily: GAD domain
50 d1xrsa_
not modelled
9.1
11
Fold: TIM beta/alpha-barrelSuperfamily: Cobalamin (vitamin B12)-dependent enzymesFamily: D-lysine 5,6-aminomutase alpha subunit, KamD
51 c3t98A_
not modelled
9.1
24
PDB header: protein transportChain: A: PDB Molecule: nuclear pore complex protein nup54;PDBTitle: molecular architecture of the transport channel of the nuclear pore2 complex: nup54/nup58
52 c6iyaD_
not modelled
8.8
19
PDB header: antitoxinChain: D: PDB Molecule: transcriptional regulator copg family;PDBTitle: structure of the dna binding domain of antitoxin copaso
53 c1zw2B_
not modelled
8.4
33
PDB header: protein bindingChain: B: PDB Molecule: talin;PDBTitle: vinculin head (0-258) in complex with the talin rod2 residues 2345-2369
54 c2gpyB_
not modelled
8.0
14
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: o-methyltransferase;PDBTitle: crystal structure of putative o-methyltransferase from bacillus2 halodurans
55 c2ebjB_
not modelled
7.6
40
PDB header: hydrolaseChain: B: PDB Molecule: pyrrolidone carboxyl peptidase;PDBTitle: crystal structure of pyrrolidone carboxyl peptidase from thermus2 thermophilus
56 d1ltqa1
not modelled
7.6
0
Fold: HAD-likeSuperfamily: HAD-likeFamily: phosphatase domain of polynucleotide kinase
57 c4d8jD_
not modelled
7.5
21
PDB header: dna binding proteinChain: D: PDB Molecule: macrodomain ter protein;PDBTitle: structure of e. coli matp-mats complex
58 c4hv0B_
not modelled
7.0
13
PDB header: transcription, viral proteinChain: B: PDB Molecule: avtr;PDBTitle: structure and function of avtr, a novel transcriptional regulator from2 a hyperthermophilic archaeal lipothrixvirus
59 d1p94a_
not modelled
5.8
16
Fold: Ribbon-helix-helixSuperfamily: Ribbon-helix-helixFamily: CopG-like
60 d1a4ia2
not modelled
5.7
19
Fold: Aminoacid dehydrogenase-like, N-terminal domainSuperfamily: Aminoacid dehydrogenase-like, N-terminal domainFamily: Tetrahydrofolate dehydrogenase/cyclohydrolase
61 c3kxeD_
not modelled
5.7
15
PDB header: protein bindingChain: D: PDB Molecule: antitoxin protein pard-1;PDBTitle: a conserved mode of protein recognition and binding in a2 pard-pare toxin-antitoxin complex
62 c3abqA_
not modelled
5.6
19
PDB header: lyaseChain: A: PDB Molecule: ethanolamine ammonia-lyase heavy chain;PDBTitle: crystal structure of ethanolamine ammonia-lyase from escherichia coli2 complexed with cn-cbl and 2-amino-1-propanol
63 c4xc8B_
not modelled
5.4
17
PDB header: isomeraseChain: B: PDB Molecule: isobutyryl-coa mutase fused;PDBTitle: isobutyryl-coa mutase fused with bound butyryl-coa, gdp, and mg and2 without cobalamin (apo-icmf/gdp)
64 c6dy3G_
not modelled
5.4
20
PDB header: hydrolaseChain: G: PDB Molecule: n-acylethanolamine-hydrolyzing acid amidase alpha-subunit;PDBTitle: caenorhabditis elegans n-acylethanolamine-hydrolyzing acid amidase2 (naaa) ortholog
65 c6dy1A_
not modelled
5.4
4
PDB header: hydrolaseChain: A: PDB Molecule: n-acylethanolamine acid amidase alpha-subunit;PDBTitle: rabbit n-acylethanolamine-hydrolyzing acid amidase (naaa) with fatty2 acid (myristate), in presence of triton x-100