| 1 | c3us8A_
|
|
 |
100.0 |
72 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:isocitrate dehydrogenase [nadp];
PDBTitle: crystal structure of an isocitrate dehydrogenase from sinorhizobium2 meliloti 1021
|
|
|
|
| 2 | c2qfyE_
|
|
 |
100.0 |
62 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:isocitrate dehydrogenase [nadp];
PDBTitle: crystal structure of saccharomyces cerevesiae mitochondrial nadp(+)-2 dependent isocitrate dehydrogenase in complex with a-ketoglutarate
|
|
|
|
| 3 | d1t0la_
|
|
 |
100.0 |
66 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
|
|
|
| 4 | d1lwda_
|
|
 |
100.0 |
66 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
|
|
|
| 5 | c2uxqB_
|
|
 |
100.0 |
51 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:isocitrate dehydrogenase native;
PDBTitle: isocitrate dehydrogenase from the psychrophilic bacterium2 desulfotalea psychrophila: biochemical properties and3 crystal structure analysis
|
|
|
|
| 6 | c1zorB_
|
|
 |
100.0 |
55 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:isocitrate dehydrogenase;
PDBTitle: isocitrate dehydrogenase from the hyperthermophile thermotoga maritima
|
|
|
|
| 7 | c4aoyD_
|
|
 |
100.0 |
53 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:isocitrate dehydrogenase [nadp];
PDBTitle: open ctidh. the complex structures of isocitrate dehydrogenase from2 clostridium thermocellum and desulfotalea psychrophila, support a new3 active site locking mechanism
|
|
|
|
| 8 | d1hqsa_
|
|
 |
100.0 |
23 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
|
|
|
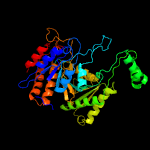
| 9 | c2d4vD_
|
|
 |
100.0 |
24 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:isocitrate dehydrogenase;
PDBTitle: crystal structure of nad dependent isocitrate dehydrogenase2 from acidithiobacillus thiooxidans
|
|
|
|
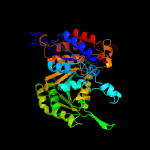
| 10 | c2d1cB_
|
|
 |
100.0 |
23 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:isocitrate dehydrogenase;
PDBTitle: crystal structure of tt0538 protein from thermus thermophilus hb8
|
|
|
|
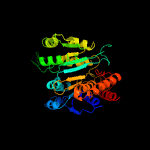
| 11 | c3fmxX_
|
|
 |
100.0 |
17 |
PDB header:oxidoreductase
Chain: X: PDB Molecule:tartrate dehydrogenase/decarboxylase;
PDBTitle: crystal structure of tartrate dehydrogenase from pseudomonas putida2 complexed with nadh
|
|
|
|
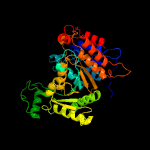
| 12 | d1pb1a_
|
|
 |
100.0 |
22 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
|
|
|
| 13 | c2e0cA_
|
|
 |
100.0 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:409aa long hypothetical nadp-dependent isocitrate
PDBTitle: crystal structure of isocitrate dehydrogenase from sulfolobus tokodaii2 strain7 at 2.0 a resolution
|
|
|
|
| 14 | d1v53a1
|
|
 |
100.0 |
19 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
|
|
|
| 15 | d1cnza_
|
|
 |
100.0 |
20 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
|
|
|
| 16 | d1cm7a_
|
|
 |
100.0 |
19 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
|
|
|
| 17 | c3uduG_
|
|
 |
100.0 |
22 |
PDB header:oxidoreductase
Chain: G: PDB Molecule:3-isopropylmalate dehydrogenase;
PDBTitle: crystal structure of putative 3-isopropylmalate dehydrogenase from2 campylobacter jejuni
|
|
|
|
| 18 | c3u1hA_
|
|
 |
100.0 |
23 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:3-isopropylmalate dehydrogenase;
PDBTitle: crystal structure of ipmdh from the last common ancestor of bacillus
|
|
|
|
| 19 | d1vlca_
|
|
 |
100.0 |
22 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
|
|
|
| 20 | c3vl3A_
|
|
 |
100.0 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:3-isopropylmalate dehydrogenase;
PDBTitle: 3-isopropylmalate dehydrogenase from shewanella oneidensis mr-1 at 3402 mpa
|
|
|
|
| 21 | d1wpwa_ |
|
not modelled |
100.0 |
20 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
|
|
| 22 | d1xaca_ |
|
not modelled |
100.0 |
21 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
|
|
| 23 | c3ty3A_ |
|
not modelled |
100.0 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:probable homoisocitrate dehydrogenase;
PDBTitle: crystal structure of homoisocitrate dehydrogenase from2 schizosaccharomyces pombe bound to glycyl-glycyl-glycine
|
|
|
| 24 | c4iwhA_ |
|
not modelled |
100.0 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:3-isopropylmalate dehydrogenase;
PDBTitle: crystal structure of a 3-isopropylmalate dehydrogenase from2 burkholderia pseudomallei
|
|
|
| 25 | d1g2ua_ |
|
not modelled |
100.0 |
21 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
|
|
| 26 | c3r8wC_ |
|
not modelled |
100.0 |
17 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:3-isopropylmalate dehydrogenase 2, chloroplastic;
PDBTitle: structure of 3-isopropylmalate dehydrogenase isoform 2 from2 arabidopsis thaliana at 2.2 angstrom resolution
|
|
|
| 27 | c1x0lB_ |
|
not modelled |
100.0 |
21 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:homoisocitrate dehydrogenase;
PDBTitle: crystal structure of tetrameric homoisocitrate dehydrogenase from an2 extreme thermophile, thermus thermophilus
|
|
|
| 28 | c1tyoA_ |
|
not modelled |
100.0 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:isocitrate dehydrogenase;
PDBTitle: isocitrate dehydrogenase from the hyperthermophile aeropyrum pernix in2 complex with etheno-nadp
|
|
|
| 29 | d1a05a_ |
|
not modelled |
100.0 |
20 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
|
|
| 30 | d1w0da_ |
|
not modelled |
100.0 |
20 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
|
|
| 31 | c5hn6A_ |
|
not modelled |
100.0 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:homoisocitrate dehydrogenase;
PDBTitle: crystal structure of beta-decarboxylating dehydrogenase (tk0280) from2 thermococcus kodakarensis complexed with mn and 3-isopropylmalate
|
|
|
| 32 | c5grhA_ |
|
not modelled |
100.0 |
24 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:isocitrate dehydrogenase [nad] subunit alpha,
PDBTitle: crystal structure of the alpha gamma heterodimer of human idh3 in2 complex with mg(2+)
|
|
|
| 33 | c3blxL_ |
|
not modelled |
100.0 |
28 |
PDB header:oxidoreductase
Chain: L: PDB Molecule:isocitrate dehydrogenase [nad] subunit 2;
PDBTitle: yeast isocitrate dehydrogenase (apo form)
|
|
|
| 34 | c5grhB_ |
|
not modelled |
100.0 |
21 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:isocitrate dehydrogenase [nad] subunit gamma,
PDBTitle: crystal structure of the alpha gamma heterodimer of human idh3 in2 complex with mg(2+)
|
|
|
| 35 | c3blxM_ |
|
not modelled |
100.0 |
21 |
PDB header:oxidoreductase
Chain: M: PDB Molecule:isocitrate dehydrogenase [nad] subunit 1;
PDBTitle: yeast isocitrate dehydrogenase (apo form)
|
|
|
| 36 | c2iv0A_ |
|
not modelled |
100.0 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:isocitrate dehydrogenase;
PDBTitle: thermal stability of isocitrate dehydrogenase from2 archaeoglobus fulgidus studied by crystal structure3 analysis and engineering of chimers
|
|
|
| 37 | c6g3uA_ |
|
not modelled |
97.9 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:isocitrate dehydrogenase;
PDBTitle: structure of pseudomonas aeruginosa isocitrate dehydrogenase, idh
|
|
|
| 38 | d1itwa_ |
|
not modelled |
97.8 |
16 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Monomeric isocitrate dehydrogenase |
|
|
| 39 | c2b0tA_ |
|
not modelled |
97.6 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nadp isocitrate dehydrogenase;
PDBTitle: structure of monomeric nadp isocitrate dehydrogenase
|
|
|
| 40 | d1r8ka_ |
|
not modelled |
96.1 |
16 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:PdxA-like |
|
|
| 41 | c4jqpA_ |
|
not modelled |
95.6 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:4-hydroxythreonine-4-phosphate dehydrogenase;
PDBTitle: x-ray crystal structure of a 4-hydroxythreonine-4-phosphate2 dehydrogenase from burkholderia phymatum
|
|
|
| 42 | c2hi1A_ |
|
not modelled |
94.6 |
21 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:4-hydroxythreonine-4-phosphate dehydrogenase 2;
PDBTitle: the structure of a putative 4-hydroxythreonine-4-phosphate2 dehydrogenase from salmonella typhimurium.
|
|
|
| 43 | d1ptma_ |
|
not modelled |
94.3 |
16 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:PdxA-like |
|
|
| 44 | c4atyA_ |
|
not modelled |
94.1 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:terephthalate 1,2-cis-dihydrodiol dehydrogenase;
PDBTitle: crystal structure of a terephthalate 1,2-cis-2 dihydrodioldehydrogenase from burkholderia xenovorans3 lb400
|
|
|
| 45 | c3tsnD_ |
|
not modelled |
93.3 |
16 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:4-hydroxythreonine-4-phosphate dehydrogenase;
PDBTitle: 4-hydroxythreonine-4-phosphate dehydrogenase from campylobacter jejuni
|
|
|
| 46 | c1yxoB_ |
|
not modelled |
88.5 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:4-hydroxythreonine-4-phosphate dehydrogenase 1;
PDBTitle: crystal structure of pyridoxal phosphate biosynthetic protein pdxa2 pa0593
|
|
|
| 47 | c5ahwC_ |
|
not modelled |
24.1 |
9 |
PDB header:signaling protein
Chain: C: PDB Molecule:universal stress protein;
PDBTitle: crystal structure of universal stress protein msmeg_3811 in2 complex with camp
|
|
|
| 48 | c1p84E_ |
|
not modelled |
21.9 |
18 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:ubiquinol-cytochrome c reductase iron-sulfur subunit;
PDBTitle: hdbt inhibited yeast cytochrome bc1 complex
|
|
|
| 49 | d1kl7a_ |
|
not modelled |
21.9 |
17 |
Fold:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family:Tryptophan synthase beta subunit-like PLP-dependent enzymes |
|
|
| 50 | c3fh0A_ |
|
not modelled |
21.8 |
12 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative universal stress protein kpn_01444;
PDBTitle: crystal structure of putative universal stress protein kpn_01444 -2 atpase
|
|
|
| 51 | c2fyuE_ |
|
not modelled |
20.6 |
18 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:ubiquinol-cytochrome c reductase iron-sulfur subunit,
PDBTitle: crystal structure of bovine heart mitochondrial bc1 with jg1442 inhibitor
|
|
|
| 52 | c5i0cA_ |
|
not modelled |
15.6 |
11 |
PDB header:hydrolase
Chain: A: PDB Molecule:uncharacterized protein yjdj;
PDBTitle: crystal structure of predicted acyltransferase yjdj with acyl-coa n-2 acyltransferase domain from escherichia coli str. k-12
|
|
|
| 53 | c2q62A_ |
|
not modelled |
12.4 |
13 |
PDB header:flavoprotein
Chain: A: PDB Molecule:arsh;
PDBTitle: crystal structure of arsh from sinorhizobium meliloti
|
|
|
| 54 | c1nvpD_ |
|
not modelled |
11.8 |
23 |
PDB header:transcription/dna
Chain: D: PDB Molecule:transcription initiation factor iia gamma chain;
PDBTitle: human tfiia/tbp/dna complex
|
|
|
| 55 | d1a0ca_ |
|
not modelled |
11.7 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:Xylose isomerase |
|
|
| 56 | d2ffea1 |
|
not modelled |
11.6 |
11 |
Fold:CofD-like
Superfamily:CofD-like
Family:CofD-like |
|
|
| 57 | d1riea_ |
|
not modelled |
11.5 |
18 |
Fold:ISP domain
Superfamily:ISP domain
Family:Rieske iron-sulfur protein (ISP) |
|
|
| 58 | c2dumD_ |
|
not modelled |
11.3 |
18 |
PDB header:structural genomics, unknown function
Chain: D: PDB Molecule:hypothetical protein ph0823;
PDBTitle: crystal structure of hypothetical protein, ph0823
|
|
|
| 59 | c4xkmB_ |
|
not modelled |
11.2 |
20 |
PDB header:isomerase
Chain: B: PDB Molecule:xylose isomerase;
PDBTitle: crystal structure of xylose isomerase from an human intestinal tract2 microbe bacteroides thetaiotaomicron
|
|
|
| 60 | d1ofda1 |
|
not modelled |
10.7 |
17 |
Fold:Single-stranded right-handed beta-helix
Superfamily:Alpha subunit of glutamate synthase, C-terminal domain
Family:Alpha subunit of glutamate synthase, C-terminal domain |
|
|
| 61 | d1u84a_ |
|
not modelled |
10.4 |
20 |
Fold:YugE-like
Superfamily:YugE-like
Family:YugE-like |
|
|
| 62 | d1mn3a_ |
|
not modelled |
10.1 |
9 |
Fold:RuvA C-terminal domain-like
Superfamily:UBA-like
Family:CUE domain |
|
|
| 63 | c5fjsB_ |
|
not modelled |
10.1 |
7 |
PDB header:hydrolase
Chain: B: PDB Molecule:glucosylceramidase;
PDBTitle: bacterial beta-glucosidase reveals the structural and functional2 basis of genetic defects in human glucocerebrosidase 2 (gba2)3 disorders
|
|
|
| 64 | c3wkxA_ |
|
not modelled |
10.1 |
21 |
PDB header:hydrolase
Chain: A: PDB Molecule:non-reducing end beta-l-arabinofuranosidase;
PDBTitle: crystal structure of gh127 beta-l-arabinofuranosidase hypba1 from2 bifidobacterium longum arabinose complex form
|
|
|
| 65 | d2o8ra3 |
|
not modelled |
10.0 |
8 |
Fold:Phospholipase D/nuclease
Superfamily:Phospholipase D/nuclease
Family:Polyphosphate kinase C-terminal domain |
|
|
| 66 | d1xmta_ |
|
not modelled |
9.5 |
11 |
Fold:Acyl-CoA N-acyltransferases (Nat)
Superfamily:Acyl-CoA N-acyltransferases (Nat)
Family:N-acetyl transferase, NAT |
|
|
| 67 | c2yu3A_ |
|
not modelled |
9.4 |
13 |
PDB header:transcription
Chain: A: PDB Molecule:dna-directed rna polymerase iii 39 kda
PDBTitle: solution structure of the domain swapped wingedhelix in dna-2 directed rna polymerase iii 39 kda polypeptide
|
|
|
| 68 | c2hb0B_ |
|
not modelled |
9.4 |
12 |
PDB header:cell adhesion
Chain: B: PDB Molecule:cfa/i fimbrial subunit e;
PDBTitle: crystal structure of cfae, the adhesive subunit of cfa/i2 fimbria of enterotoxigenic escherichia coli
|
|
|
| 69 | c6bwsA_ |
|
not modelled |
9.3 |
10 |
PDB header:unknown function
Chain: A: PDB Molecule:glycolate utilization protein;
PDBTitle: crystal structure of efga from methylobacterium extorquens
|
|
|
| 70 | d2d9ra1 |
|
not modelled |
9.2 |
20 |
Fold:Double-split beta-barrel
Superfamily:AF2212/PG0164-like
Family:PG0164-like |
|
|
| 71 | c3rhtB_ |
|
not modelled |
9.2 |
11 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:(gatase1)-like protein;
PDBTitle: crystal structure of type 1 glutamine amidotransferase (gatase1)-like2 protein from planctomyces limnophilus
|
|
|
| 72 | d1a0ea_ |
|
not modelled |
9.0 |
10 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:Xylose isomerase |
|
|
| 73 | d2gana1 |
|
not modelled |
8.9 |
10 |
Fold:Acyl-CoA N-acyltransferases (Nat)
Superfamily:Acyl-CoA N-acyltransferases (Nat)
Family:N-acetyl transferase, NAT |
|
|
| 74 | c2pfsA_ |
|
not modelled |
8.8 |
9 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:universal stress protein;
PDBTitle: crystal structure of universal stress protein from nitrosomonas2 europaea
|
|
|
| 75 | c2j3zA_ |
|
not modelled |
8.6 |
16 |
PDB header:toxin
Chain: A: PDB Molecule:c2 toxin component i;
PDBTitle: crystal structure of the enzymatic component c2-i of the c2-toxin from2 clostridium botulinum at ph 6.1
|
|
|
| 76 | c6ex6A_ |
|
not modelled |
8.6 |
21 |
PDB header:hydrolase
Chain: A: PDB Molecule:six-hairpin glycosidase;
PDBTitle: the gh127, beta-arabinofuranosidase, bt3674
|
|
|
| 77 | c1nh2D_ |
|
not modelled |
8.6 |
19 |
PDB header:transcription/dna
Chain: D: PDB Molecule:transcription initiation factor iia small chain;
PDBTitle: crystal structure of a yeast tfiia/tbp/dna complex
|
|
|
| 78 | c6dxpD_ |
|
not modelled |
8.4 |
16 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:fmn-dependent nadh-azoreductase;
PDBTitle: the crystal structure of an fmn-dependent nadh-azoreductase from2 klebsiella pneumoniae
|
|
|
| 79 | c2ozhA_ |
|
not modelled |
8.3 |
8 |
PDB header:transferase
Chain: A: PDB Molecule:hypothetical protein xcc2953;
PDBTitle: crystal structure of a putative acetyltransferase belonging to the2 gnat family (xcc2953) from xanthomonas campestris pv. campestris at3 1.40 a resolution
|
|
|
| 80 | c4wnyA_ |
|
not modelled |
8.2 |
9 |
PDB header:signaling protein
Chain: A: PDB Molecule:universal stress protein;
PDBTitle: crystal structure of a protein from the universal stress protein2 family from burkholderia pseudomallei
|
|
|
| 81 | c5m1hA_ |
|
not modelled |
8.1 |
18 |
PDB header:viral protein
Chain: A: PDB Molecule:gag protein;
PDBTitle: structure of a spumaretrovirus gag central domain reveals an ancient2 retroviral capsid
|
|
|
| 82 | c3f83A_ |
|
not modelled |
7.9 |
12 |
PDB header:cell adhesion
Chain: A: PDB Molecule:fusion of the minor pilin cfae and major pilin cfab;
PDBTitle: structure of fusion complex of the minor pilin cfae and major pilin2 cfab of cfa/i pili from etec e. coli
|
|
|
| 83 | c5dniB_ |
|
not modelled |
7.9 |
13 |
PDB header:lyase
Chain: B: PDB Molecule:putative l(+)-tartrate dehydratase subunit beta;
PDBTitle: crystal structure of methanocaldococcus jannaschii fumarate hydratase2 beta subunit
|
|
|
| 84 | d1q77a_ |
|
not modelled |
7.8 |
3 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:Universal stress protein-like |
|
|
| 85 | d1qasa3 |
|
not modelled |
7.7 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:PLC-like phosphodiesterases
Family:Mammalian PLC |
|
|
| 86 | d1t9ba1 |
|
not modelled |
7.4 |
11 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Pyruvate oxidase and decarboxylase, middle domain |
|
|
| 87 | c2p0yA_ |
|
not modelled |
7.3 |
17 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein lp_0780;
PDBTitle: crystal structure of q88yi3_lacpl from lactobacillus plantarum.2 northeast structural genomics consortium target lpr6
|
|
|
| 88 | c4pc4B_ |
|
not modelled |
7.3 |
7 |
PDB header:lipid binding protein
Chain: B: PDB Molecule:30k lipoprotein;
PDBTitle: bombyx mori lipoprotein 6
|
|
|
| 89 | d1jmva_ |
|
not modelled |
7.3 |
6 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:Universal stress protein-like |
|
|
| 90 | c2pt7G_ |
|
not modelled |
7.1 |
11 |
PDB header:hydrolase/protein binding
Chain: G: PDB Molecule:hypothetical protein;
PDBTitle: crystal structure of cag virb11 (hp0525) and an inhibitory protein2 (hp1451)
|
|
|
| 91 | c2jaxA_ |
|
not modelled |
7.1 |
12 |
PDB header:protein binding
Chain: A: PDB Molecule:hypothetical protein tb31.7;
PDBTitle: universal stress protein rv2623 from mycobaterium2 tuberculosis
|
|
|
| 92 | c3pubA_ |
|
not modelled |
7.0 |
11 |
PDB header:unknown function
Chain: A: PDB Molecule:30kda protein;
PDBTitle: crystal structure of the bombyx mori low molecular weight lipoprotein2 7 (bmlp7)
|
|
|
| 93 | c2kvoA_ |
|
not modelled |
6.9 |
13 |
PDB header:photosynthesis
Chain: A: PDB Molecule:photosystem ii reaction center psb28 protein;
PDBTitle: solution nmr structure of photosystem ii reaction center psb28 protein2 from synechocystis sp.(strain pcc 6803), northeast structural3 genomics consortium target sgr171
|
|
|
| 94 | d1tj1a2 |
|
not modelled |
6.9 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:FAD-linked oxidoreductase
Family:Proline dehydrohenase domain of bifunctional PutA protein |
|
|
| 95 | d1i27a_ |
|
not modelled |
6.9 |
28 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:C-terminal domain of the rap74 subunit of TFIIF |
|
|
| 96 | c3dddA_ |
|
not modelled |
6.8 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:putative acetyltransferase;
PDBTitle: crystal structure of a putative acetyltransferase (np_142035.1) from2 pyrococcus horikoshii at 2.25 a resolution
|
|
|
| 97 | c3zpnD_ |
|
not modelled |
6.7 |
11 |
PDB header:photosynthesis
Chain: D: PDB Molecule:photosystem ii reaction center psb28 protein;
PDBTitle: structure of psb28
|
|
|
| 98 | c2fynO_ |
|
not modelled |
6.6 |
16 |
PDB header:oxidoreductase
Chain: O: PDB Molecule:ubiquinol-cytochrome c reductase iron-sulfur
PDBTitle: crystal structure analysis of the double mutant rhodobacter2 sphaeroides bc1 complex
|
|
|
| 99 | c6hhnA_ |
|
not modelled |
6.6 |
21 |
PDB header:isomerase
Chain: A: PDB Molecule:l-rhamnose mutarotase;
PDBTitle: crystal structure of l-rhamnose mutarotase fa22100 from formosa2 agariphila
|
|
|











































































































































































































































































