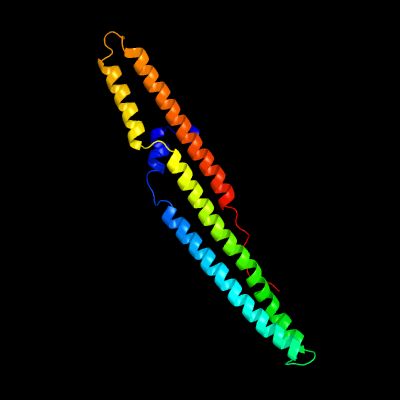
1 c5xfsB_
100.0
52
PDB header: protein transportChain: B: PDB Molecule: ppe family protein ppe15;PDBTitle: crystal structure of pe8-ppe15 in complex with espg5 from m.2 tuberculosis
2 c2g38B_
100.0
33
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: ppe family protein;PDBTitle: a pe/ppe protein complex from mycobacterium tuberculosis
3 d2g38b1
100.0
33
Fold: Ferritin-likeSuperfamily: PE/PPE dimer-likeFamily: PPE
4 c4xy3A_
100.0
19
PDB header: protein transportChain: A: PDB Molecule: esx-1 secretion-associated protein espb;PDBTitle: structure of esx-1 secreted protein espb
5 c4wj2A_
97.0
19
PDB header: unknown functionChain: A: PDB Molecule: antigen mtb48;PDBTitle: mycobacterial protein
6 c2vs0B_
96.6
10
PDB header: cell invasionChain: B: PDB Molecule: virulence factor esxa;PDBTitle: structural analysis of homodimeric staphylococcal aureus2 virulence factor esxa
7 c4iogD_
96.4
14
PDB header: unknown functionChain: D: PDB Molecule: secreted protein esxb;PDBTitle: the crystal structure of a secreted protein esxb (wild-type, in p212 space group) from bacillus anthracis str. sterne
8 c3gvmA_
96.0
11
PDB header: viral proteinChain: A: PDB Molecule: putative uncharacterized protein sag1039;PDBTitle: structure of the homodimeric wxg-100 family protein from streptococcus2 agalactiae
9 c3zbhC_
95.5
15
PDB header: unknown functionChain: C: PDB Molecule: esxa;PDBTitle: geobacillus thermodenitrificans esxa crystal form i
10 d1wa8a1
94.0
18
Fold: Ferritin-likeSuperfamily: EsxAB dimer-likeFamily: ESAT-6 like
11 c3jywF_
82.2
54
PDB header: ribosomeChain: F: PDB Molecule: 60s ribosomal protein l7(a);PDBTitle: structure of the 60s proteins for eukaryotic ribosome based on cryo-em2 map of thermomyces lanuginosus ribosome at 8.9a resolution
12 c5kf6B_
80.7
18
PDB header: oxidoreductaseChain: B: PDB Molecule: bifunctional protein puta;PDBTitle: structure of proline utilization a from sinorhizobium meliloti2 complexed with l-tetrahydrofuroic acid and nad+ in space group p21
13 c4i0xA_
80.2
26
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: esat-6-like protein mab_3112;PDBTitle: crystal structure of the mycobacterum abscessus esxef (mab_3112-2 mab_3113) complex
14 c2vsnB_
78.9
16
PDB header: transferaseChain: B: PDB Molecule: xcogt;PDBTitle: structure and topological arrangement of an o-glcnac2 transferase homolog: insight into molecular control of3 intracellular glycosylation
15 c5nxhB_
77.1
15
PDB header: viral proteinChain: B: PDB Molecule: long-tail fiber proximal subunit;PDBTitle: crystal structure of the carboxy-terminal region of the bacteriophage2 t4 proximal long tail fibre protein gp34, residues 744-1289 at 2.93 angstrom resolution
16 c2qqpG_
75.2
16
PDB header: virusChain: G: PDB Molecule: large capsid protein;PDBTitle: crystal structure of authentic providence virus
17 d1wa8b1
74.2
20
Fold: Ferritin-likeSuperfamily: EsxAB dimer-likeFamily: ESAT-6 like
18 c4lwsA_
73.6
21
PDB header: unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: esxa : esxb (semet) hetero-dimer from thermomonospora curvata
19 c4f9iA_
72.1
17
PDB header: oxidoreductaseChain: A: PDB Molecule: proline dehydrogenase/delta-1-pyrroline-5-carboxylatePDBTitle: crystal structure of proline utilization a (puta) from geobacter2 sulfurreducens pca
20 d1rp4a_
70.6
23
Fold: ERO1-likeSuperfamily: ERO1-likeFamily: ERO1-like
21 c3pe3D_
not modelled
64.8
18
PDB header: transferaseChain: D: PDB Molecule: udp-n-acetylglucosamine--peptide n-PDBTitle: structure of human o-glcnac transferase and its complex with a peptide2 substrate
22 c3ahrA_
not modelled
63.0
18
PDB header: oxidoreductaseChain: A: PDB Molecule: ero1-like protein alpha;PDBTitle: inactive human ero1
23 c2vakF_
not modelled
59.6
19
PDB header: viral proteinChain: F: PDB Molecule: sigma a;PDBTitle: crystal structure of the avian reovirus inner capsid protein sigmaa
24 c3at7B_
not modelled
57.1
17
PDB header: structural proteinChain: B: PDB Molecule: alginate-binding flagellin;PDBTitle: crystal structure of bacterial cell-surface alginate-binding protein2 algp7
25 c4lwsB_
not modelled
55.3
15
PDB header: unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: esxa : esxb (semet) hetero-dimer from thermomonospora curvata
26 c3hazA_
not modelled
53.3
28
PDB header: oxidoreductaseChain: A: PDB Molecule: proline dehydrogenase;PDBTitle: crystal structure of bifunctional proline utilization a2 (puta) protein
27 d1ncfb3
not modelled
52.8
38
Fold: TNF receptor-likeSuperfamily: TNF receptor-likeFamily: TNF receptor-like
28 d1exta3
not modelled
52.5
38
Fold: TNF receptor-likeSuperfamily: TNF receptor-likeFamily: TNF receptor-like
29 d2fr1a1
not modelled
51.4
16
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Tyrosine-dependent oxidoreductases
30 d1ui5a2
not modelled
51.3
16
Fold: Tetracyclin repressor-like, C-terminal domainSuperfamily: Tetracyclin repressor-like, C-terminal domainFamily: Tetracyclin repressor-like, C-terminal domain
31 c2wybA_
not modelled
50.8
12
PDB header: hydrolaseChain: A: PDB Molecule: acyl-homoserine lactone acylase pvdq subunitPDBTitle: the quorum quenching n-acyl homoserine lactone acylase pvdq2 with a covalently bound dodecanoic acid
32 c5djsA_
not modelled
49.6
23
PDB header: transferaseChain: A: PDB Molecule: tetratricopeptide tpr_2 repeat protein;PDBTitle: thermobaculum terrenum o-glcnac transferase mutant - k341m
33 c3lhoA_
not modelled
49.3
50
PDB header: hydrolaseChain: A: PDB Molecule: putative hydrolase;PDBTitle: crystal structure of putative hydrolase (yp_751971.1) from shewanella2 frigidimarina ncimb 400 at 1.80 a resolution
34 c4e4gF_
not modelled
49.2
14
PDB header: oxidoreductaseChain: F: PDB Molecule: methylmalonate-semialdehyde dehydrogenase;PDBTitle: crystal structure of putative methylmalonate-semialdehyde2 dehydrogenase from sinorhizobium meliloti 1021
35 c2vroB_
not modelled
48.6
22
PDB header: oxidoreductaseChain: B: PDB Molecule: aldehyde dehydrogenase;PDBTitle: crystal structure of aldehyde dehydrogenase from2 burkholderia xenovorans lb400
36 c2kg7B_
not modelled
48.0
20
PDB header: unknown functionChain: B: PDB Molecule: esat-6-like protein esxh;PDBTitle: structure and features of the complex formed by the tuberculosis2 virulence factors rv0287 and rv0288
37 d3cx5d1
not modelled
47.3
25
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: Cytochrome bc1 domain
38 c2ve5H_
not modelled
46.6
18
PDB header: oxidoreductaseChain: H: PDB Molecule: betaine aldehyde dehydrogenase;PDBTitle: crystallographic structure of betaine aldehyde2 dehydrogenase from pseudomonas aeruginosa
39 d1ppjd1
not modelled
45.9
20
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: Cytochrome bc1 domain
40 c4bgoA_
not modelled
45.6
12
PDB header: hydrolaseChain: A: PDB Molecule: efem m75 peptidase;PDBTitle: structural and functional role of the imelysin-like protein2 efem from pseudomonas syringae pv. syringae and3 implications in bacterial iron transport
41 d1xkna_
not modelled
44.9
18
Fold: Pentein, beta/alpha-propellerSuperfamily: PenteinFamily: Porphyromonas-type peptidylarginine deiminase
42 c3ed6B_
not modelled
44.9
14
PDB header: oxidoreductaseChain: B: PDB Molecule: betaine aldehyde dehydrogenase;PDBTitle: 1.7 angstrom resolution crystal structure of betaine aldehyde2 dehydrogenase (betb) from staphylococcus aureus
43 d1oy0a_
not modelled
43.7
22
Fold: TIM beta/alpha-barrelSuperfamily: Phosphoenolpyruvate/pyruvate domainFamily: Ketopantoate hydroxymethyltransferase PanB
44 c4h7nA_
not modelled
43.4
18
PDB header: oxidoreductaseChain: A: PDB Molecule: aldehyde dehydrogenase;PDBTitle: the structure of putative aldehyde dehydrogenase puta from anabaena2 variabilis.
45 c4o5hD_
not modelled
42.7
21
PDB header: oxidoreductaseChain: D: PDB Molecule: phenylacetaldehyde dehydrogenase;PDBTitle: x-ray crystal structure of a putative phenylacetaldehyde dehydrogenase2 from burkholderia cenocepacia
46 c2jg7G_
not modelled
40.7
17
PDB header: oxidoreductaseChain: G: PDB Molecule: antiquitin;PDBTitle: crystal structure of seabream antiquitin and elucidation of2 its substrate specificity
47 c3qx3B_
not modelled
40.2
31
PDB header: isomerase/dna/isomerase inhibitorChain: B: PDB Molecule: dna topoisomerase 2-beta;PDBTitle: human topoisomerase iibeta in complex with dna and etoposide
48 c2mu3A_
not modelled
38.0
17
PDB header: structural proteinChain: A: PDB Molecule: aciniform spidroin 1;PDBTitle: spider wrapping silk fibre architecture arising from its modular2 soluble protein precursor
49 c1qcrD_
not modelled
37.3
20
PDB header: oxidoreductaseChain: D: PDB Molecule: ubiquinol cytochrome c oxidoreductase;PDBTitle: crystal structure of bovine mitochondrial cytochrome bc12 complex, alpha carbon atoms only
50 c4nl6C_
not modelled
37.3
71
PDB header: splicingChain: C: PDB Molecule: survival motor neuron protein;PDBTitle: structure of the full-length form of the protein smn found in healthy2 patients
51 c4fm9A_
not modelled
36.7
27
PDB header: isomerase/dnaChain: A: PDB Molecule: dna topoisomerase 2-alpha;PDBTitle: human topoisomerase ii alpha bound to dna
52 c5fhzF_
not modelled
36.3
14
PDB header: oxidoreductaseChain: F: PDB Molecule: aldehyde dehydrogenase family 1 member a3;PDBTitle: human aldehyde dehydrogenase 1a3 complexed with nad(+) and retinoic2 acid
53 c4lrvL_
not modelled
35.5
40
PDB header: dna binding proteinChain: L: PDB Molecule: dna sulfur modification protein dnde;PDBTitle: crystal structure of dnde from escherichia coli b7a involved in dna2 phosphorothioation modification
54 c1zrtD_
not modelled
35.1
30
PDB header: oxidoreductase/metal transportChain: D: PDB Molecule: cytochrome c1;PDBTitle: rhodobacter capsulatus cytochrome bc1 complex with2 stigmatellin bound
55 d1tpla_
not modelled
35.1
13
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: Beta-eliminating lyases
56 c4q7oA_
not modelled
32.7
32
PDB header: immune systemChain: A: PDB Molecule: immunity protein;PDBTitle: the crystal structure of an immunity protein nmb0503 from neisseria2 meningitidis mc58
57 c3cwbQ_
not modelled
32.6
20
PDB header: oxidoreductaseChain: Q: PDB Molecule: mitochondrial cytochrome c1, heme protein;PDBTitle: chicken cytochrome bc1 complex inhibited by an iodinated analogue of2 the polyketide crocacin-d
58 c4rqoB_
not modelled
31.9
32
PDB header: lyaseChain: B: PDB Molecule: l-serine dehydratase;PDBTitle: crystal structure of l-serine dehydratase from legionella pneumophila
59 c2hg2A_
not modelled
31.9
19
PDB header: oxidoreductaseChain: A: PDB Molecule: aldehyde dehydrogenase a;PDBTitle: structure of lactaldehyde dehydrogenase
60 c5ujuA_
not modelled
31.7
17
PDB header: oxidoreductaseChain: A: PDB Molecule: nad-dependent aldehyde dehydrogenase;PDBTitle: crystal structure of nad-dependent aldehyde dehydrogenase from2 burkholderia multivorans
61 c3u4jB_
not modelled
31.5
20
PDB header: oxidoreductaseChain: B: PDB Molecule: nad-dependent aldehyde dehydrogenase;PDBTitle: crystal structure of nad-dependent aldehyde dehydrogenase from2 sinorhizobium meliloti
62 c6ghrF_
not modelled
30.7
59
PDB header: photosynthesisChain: F: PDB Molecule: cp12 polypeptide;PDBTitle: cyanobacterial gapdh with full-length cp12
63 d1bi9a_
not modelled
30.5
15
Fold: ALDH-likeSuperfamily: ALDH-likeFamily: ALDH-like
64 c3fpjA_
not modelled
30.3
11
PDB header: biosynthetic protein, transferaseChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: crystal structure of e81q mutant of mtnas in complex with s-2 adenosylmethionine
65 c5lzkB_
not modelled
30.2
9
PDB header: structural genomicsChain: B: PDB Molecule: protein fam83b;PDBTitle: structure of the domain of unknown function duf1669 from human fam83b
66 c1bjtA_
not modelled
30.1
23
PDB header: topoisomeraseChain: A: PDB Molecule: topoisomerase ii;PDBTitle: topoisomerase ii residues 409-1201
67 d1bjta_
not modelled
30.1
23
Fold: Type II DNA topoisomeraseSuperfamily: Type II DNA topoisomeraseFamily: Type II DNA topoisomerase
68 c3rf7A_
not modelled
29.4
12
PDB header: oxidoreductaseChain: A: PDB Molecule: iron-containing alcohol dehydrogenase;PDBTitle: crystal structure of an iron-containing alcohol dehydrogenase2 (sden_2133) from shewanella denitrificans os-217 at 2.12 a resolution
69 c4urjA_
not modelled
29.3
19
PDB header: unknown functionChain: A: PDB Molecule: protein fam83a;PDBTitle: crystal structure of human bj-tsa-9
70 d1byra_
not modelled
29.2
17
Fold: Phospholipase D/nucleaseSuperfamily: Phospholipase D/nucleaseFamily: Nuclease
71 d2a15a1
not modelled
29.1
16
Fold: Cystatin-likeSuperfamily: NTF2-likeFamily: Ketosteroid isomerase-like
72 c2yiuE_
not modelled
28.7
35
PDB header: oxidoreductaseChain: E: PDB Molecule: cytochrome c1, heme protein;PDBTitle: x-ray structure of the dimeric cytochrome bc1 complex from2 the soil bacterium paracoccus denitrificans at 2.73 angstrom resolution
73 d2gxfa1
not modelled
28.4
16
Fold: Cystatin-likeSuperfamily: NTF2-likeFamily: YybH-like
74 c4i3wC_
not modelled
28.3
21
PDB header: oxidoreductaseChain: C: PDB Molecule: aldehyde dehydrogenase (nad+);PDBTitle: structure of phosphonoacetaldehyde dehydrogenase in complex with2 gylceraldehyde-3-phosphate and cofactor nad+
75 c6fhjA_
not modelled
27.6
20
PDB header: hydrolaseChain: A: PDB Molecule: protein,protein;PDBTitle: structural dynamics and catalytic properties of a multi-modular2 xanthanase, native.
76 c3vohA_
not modelled
27.5
16
PDB header: hydrolaseChain: A: PDB Molecule: cellobiohydrolase;PDBTitle: cccel6a catalytic domain complexed with cellobiose
77 c2ordA_
not modelled
27.5
16
PDB header: transferaseChain: A: PDB Molecule: acetylornithine aminotransferase;PDBTitle: crystal structure of acetylornithine aminotransferase (ec 2.6.1.11)2 (acoat) (tm1785) from thermotoga maritima at 1.40 a resolution
78 c3q3hA_
not modelled
27.0
10
PDB header: transferaseChain: A: PDB Molecule: hmw1c-like glycosyltransferase;PDBTitle: crystal structure of the actinobacillus pleuropneumoniae hmw1c2 glycosyltransferase in complex with udp-glc
79 d1vkpa_
not modelled
27.0
31
Fold: Pentein, beta/alpha-propellerSuperfamily: PenteinFamily: Porphyromonas-type peptidylarginine deiminase
80 d1luaa2
not modelled
27.0
27
Fold: Aminoacid dehydrogenase-like, N-terminal domainSuperfamily: Aminoacid dehydrogenase-like, N-terminal domainFamily: Methylene-tetrahydromethanopterin dehydrogenase
81 c4gfhA_
not modelled
26.4
23
PDB header: isomerase/dnaChain: A: PDB Molecule: dna topoisomerase 2;PDBTitle: topoisomerase ii-dna-amppnp complex
82 c3kb4D_
not modelled
26.3
19
PDB header: structural genomics, unknown functionChain: D: PDB Molecule: alr8543 protein;PDBTitle: crystal structure of the alr8543 protein in complex with2 geranylgeranyl monophosphate and magnesium ion from nostoc sp. pcc3 7120, northeast structural genomics consortium target nsr141
83 d2ewoa1
not modelled
26.2
17
Fold: Pentein, beta/alpha-propellerSuperfamily: PenteinFamily: Porphyromonas-type peptidylarginine deiminase
84 c1p84D_
not modelled
26.0
25
PDB header: oxidoreductaseChain: D: PDB Molecule: cytochrome c1, heme protein;PDBTitle: hdbt inhibited yeast cytochrome bc1 complex
85 c2fynH_
not modelled
25.3
30
PDB header: oxidoreductaseChain: H: PDB Molecule: cytochrome c1;PDBTitle: crystal structure analysis of the double mutant rhodobacter2 sphaeroides bc1 complex
86 c5aizA_
not modelled
25.1
17
PDB header: zinc-binding proteinChain: A: PDB Molecule: zinc finger miz domain-containing protein 1;PDBTitle: the pias-like coactivator zmiz1 is a direct and selective cofactor2 of notch1 in t-cell development and leukemia
87 d1o66a_
not modelled
24.9
22
Fold: TIM beta/alpha-barrelSuperfamily: Phosphoenolpyruvate/pyruvate domainFamily: Ketopantoate hydroxymethyltransferase PanB
88 c4jzaB_
not modelled
24.9
27
PDB header: hydrolaseChain: B: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a legionella phosphoinositide phosphatase:2 insights into lipid metabolism in pathogen host interaction
89 c6dbbA_
not modelled
24.8
16
PDB header: oxidoreductaseChain: A: PDB Molecule: putative aldehyde dehydrogenase family protein;PDBTitle: crystal structure of a putative aldehyde dehydrogenase family protein2 burkholderia cenocepacia j2315 in complex with partially reduced nadh
90 d3cu3a1
not modelled
24.8
24
Fold: Cystatin-likeSuperfamily: NTF2-likeFamily: BaiE/LinA-like
91 c5azaA_
not modelled
24.6
17
PDB header: sugar binding protein, transferaseChain: A: PDB Molecule: maltose-binding periplasmic protein,oligosaccharylPDBTitle: crystal structure of mbp-saglb fusion protein with a 20-residue spacer2 in the connector helix
92 c6fhnA_
not modelled
24.6
20
PDB header: hydrolaseChain: A: PDB Molecule: protein;PDBTitle: structural dynamics and catalytic properties of a multi-modular2 xanthanase (pt derivative)
93 d1bvsa1
not modelled
24.5
36
Fold: RuvA C-terminal domain-likeSuperfamily: DNA helicase RuvA subunit, C-terminal domainFamily: DNA helicase RuvA subunit, C-terminal domain
94 c3b5oA_
not modelled
24.1
50
PDB header: oxidoreductaseChain: A: PDB Molecule: cadd-like protein of unknown function;PDBTitle: crystal structure of a cadd-like protein of unknown function2 (npun_f6505) from nostoc punctiforme pcc 73102 at 1.35 a resolution
95 d1vefa1
not modelled
24.1
14
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: GABA-aminotransferase-like
96 c2pheC_
not modelled
24.1
31
PDB header: transcriptionChain: C: PDB Molecule: alpha trans-inducing protein;PDBTitle: model for vp16 binding to pc4
97 c2wk1A_
not modelled
24.0
33
PDB header: transferaseChain: A: PDB Molecule: novp;PDBTitle: structure of the o-methyltransferase novp
98 c6ca8A_
not modelled
23.9
19
PDB header: isomeraseChain: A: PDB Molecule: dna topoisomerase 2;PDBTitle: crystal structure of plasmodium falciparum topoisomerase ii dna-2 binding, cleavage and re-ligation domain
99 c6ajrA_
not modelled
23.7
27
PDB header: dna binding proteinChain: A: PDB Molecule: uracil dna glycosylase superfamily protein;PDBTitle: complex form of uracil dna glycosylase x and uracil
100 c4ovmE_
not modelled
23.4
28
PDB header: unknown functionChain: E: PDB Molecule: uncharacterized protein sgcj;PDBTitle: crystal structure of sgcj protein from streptomyces carzinostaticus
101 d1kzqa1
not modelled
23.3
25
Fold: Cupredoxin-likeSuperfamily: Major surface antigen p30, SAG1Family: Major surface antigen p30, SAG1
102 c6iz9B_
not modelled
23.3
14
PDB header: transferaseChain: B: PDB Molecule: beta-transaminase;PDBTitle: crystal structure of the apo form of a beta-transaminase from2 mesorhizobium sp. strain luk
103 c3ifzA_
not modelled
23.0
30
PDB header: isomeraseChain: A: PDB Molecule: dna gyrase subunit a;PDBTitle: crystal structure of the first part of the mycobacterium tuberculosis2 dna gyrase reaction core: the breakage and reunion domain at 2.7 a3 resolution
104 c2k2uB_
not modelled
23.0
31
PDB header: transcriptionChain: B: PDB Molecule: alpha trans-inducing protein;PDBTitle: nmr structure of the complex between tfb1 subunit of tfiih2 and the activation domain of vp16
105 c3hx8A_
not modelled
23.0
27
PDB header: isomeraseChain: A: PDB Molecule: putative ketosteroid isomerase;PDBTitle: crystal structure of putative ketosteroid isomerase (np_103587.1) from2 mesorhizobium loti at 1.45 a resolution
106 c3r8rJ_
not modelled
22.7
18
PDB header: transferaseChain: J: PDB Molecule: transaldolase;PDBTitle: transaldolase from bacillus subtilis
107 d1nfpa_
not modelled
22.2
17
Fold: TIM beta/alpha-barrelSuperfamily: Bacterial luciferase-likeFamily: Non-fluorescent flavoprotein (luxF, FP390)
108 c5ur2C_
not modelled
22.1
19
PDB header: oxidoreductaseChain: C: PDB Molecule: bifunctional protein puta;PDBTitle: crystal structure of proline utilization a (puta) from bdellovibrio2 bacteriovorus inactivated by n-propargylglycine
109 d1jpdx2
not modelled
21.8
21
Fold: Enolase N-terminal domain-likeSuperfamily: Enolase N-terminal domain-likeFamily: Enolase N-terminal domain-like
110 d2jera1
not modelled
21.8
20
Fold: Pentein, beta/alpha-propellerSuperfamily: PenteinFamily: Porphyromonas-type peptidylarginine deiminase
111 c4yfbD_
not modelled
21.5
22
PDB header: hydrolaseChain: D: PDB Molecule: protein related to penicillin acylase;PDBTitle: structure of n-acylhomoserine lactone acylase macq in complex with2 phenylacetic acid
112 c3pefA_
not modelled
21.4
13
PDB header: oxidoreductaseChain: A: PDB Molecule: 6-phosphogluconate dehydrogenase, nad-binding;PDBTitle: crystal structure of gamma-hydroxybutyrate dehydrogenase from2 geobacter metallireducens in complex with nadp+
113 c6cboB_
not modelled
21.0
19
PDB header: transferaseChain: B: PDB Molecule: c-6' aminotransferase;PDBTitle: x-ray structure of genb1 from micromonospora echinospora in complex2 with neamine and plp (as the external aldimine)
114 c4xvzB_
not modelled
20.6
33
PDB header: transferaseChain: B: PDB Molecule: mycinamicin iii 3''-o-methyltransferase;PDBTitle: mycf mycinamicin iii 3'-o-methyltransferase in complex with mg
115 d1vpxa_
not modelled
20.4
18
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class I aldolase
116 d1joga_
not modelled
20.3
26
Fold: Four-helical up-and-down bundleSuperfamily: Nucleotidyltransferase substrate binding subunit/domainFamily: Family 1 bi-partite nucleotidyltransferase subunit
117 c3r5zB_
not modelled
20.3
22
PDB header: unknown functionChain: B: PDB Molecule: putative uncharacterized protein;PDBTitle: structure of a deazaflavin-dependent reductase from nocardia2 farcinica, with co-factor f420
118 c3omeE_
not modelled
20.3
15
PDB header: lyaseChain: E: PDB Molecule: enoyl-coa hydratase;PDBTitle: crystal structure of a probable enoyl-coa hydratase from mycobacterium2 smegmatis
119 c5ddwD_
not modelled
20.2
14
PDB header: transferaseChain: D: PDB Molecule: crmg;PDBTitle: crystal structure of aminotransferase crmg from actinoalloteichus sp.2 wh1-2216-6 in complex with the pmp external aldimine adduct with3 caerulomycin m
120 c2jerG_
not modelled
20.2
20
PDB header: hydrolaseChain: G: PDB Molecule: agmatine deiminase;PDBTitle: agmatine deiminase of enterococcus faecalis catalyzing its2 reaction.