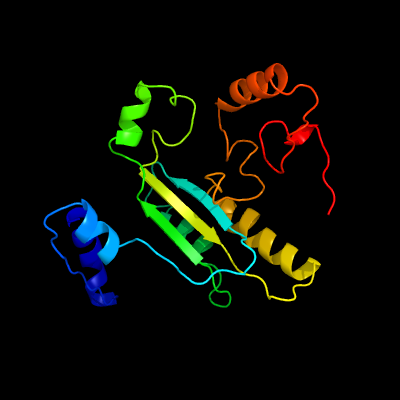
| 1 | c6f74B_
|
|
 |
99.9 |
17 |
PDB header:flavoprotein
Chain: B: PDB Molecule:alcohol oxidase;
PDBTitle: crystal structure of vao-type flavoprotein mtvao713 from2 myceliophthora thermophila c1
|
|
|
|
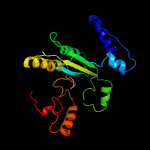
| 2 | c6f73B_
|
|
 |
99.9 |
16 |
PDB header:flavoprotein
Chain: B: PDB Molecule:mtvao615;
PDBTitle: crystal structure of vao-type flavoprotein mtvao615 at ph 5.0 from2 myceliophthora thermophila c1
|
|
|
|
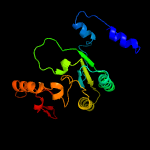
| 3 | c3popD_
|
|
 |
99.9 |
16 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:gilr oxidase;
PDBTitle: the crystal structure of gilr, an oxidoreductase that catalyzes the2 terminal step of gilvocarcin biosynthesis
|
|
|
|
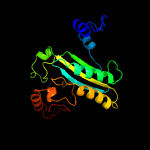
| 4 | c2wdwB_
|
|
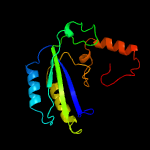 |
99.9 |
22 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative hexose oxidase;
PDBTitle: the native crystal structure of the primary hexose oxidase (2 dbv29) in antibiotic a40926 biosynthesis
|
|
|
|
| 5 | c5i1wD_
|
|
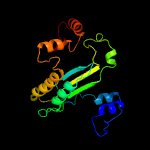 |
99.8 |
19 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:crmk;
PDBTitle: crystal structure of crmk, a flavoenzyme involved in the shunt product2 recycling mechanism in caerulomycin biosynthesis
|
|
|
|
| 6 | c2y3rC_
|
|
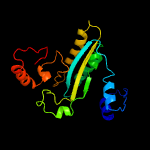 |
99.8 |
16 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:taml;
PDBTitle: structure of the tirandamycin-bound fad-dependent tirandamycin oxidase2 taml in p21 space group
|
|
|
|
| 7 | c2ipiD_
|
|
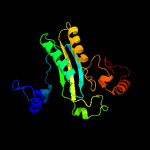 |
99.8 |
18 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:aclacinomycin oxidoreductase (aknox);
PDBTitle: crystal structure of aclacinomycin oxidoreductase
|
|
|
|
| 8 | c4ud8B_
|
|
 |
99.8 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:fad-binding and bbe domain-containing protein;
PDBTitle: atbbe15
|
|
|
|
| 9 | c6eo5A_
|
|
 |
99.8 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:ppbbe-like 1 d396n;
PDBTitle: physcomitrella patens bbe-like 1 variant d396n
|
|
|
|
| 10 | c3vteA_
|
|
 |
99.8 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:tetrahydrocannabinolic acid synthase;
PDBTitle: crystal structure of tetrahydrocannabinolic acid synthase from2 cannabis sativa
|
|
|
|
| 11 | c3tsjA_
|
|
 |
99.8 |
19 |
PDB header:allergen, oxidoreductase
Chain: A: PDB Molecule:pollen allergen phl p 4;
PDBTitle: crystal structure of phl p 4, a grass pollen allergen with glucose2 dehydrogenase activity
|
|
|
|
| 12 | c3w8wA_
|
|
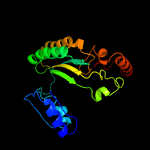 |
99.8 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative fad-dependent oxygenase encm;
PDBTitle: the crystal structure of encm
|
|
|
|
| 13 | c3d2hA_
|
|
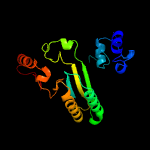 |
99.8 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:berberine bridge-forming enzyme;
PDBTitle: structure of berberine bridge enzyme from eschscholzia californica,2 monoclinic crystal form
|
|
|
|
| 14 | c5d79B_
|
|
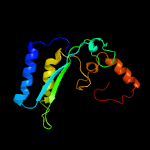 |
99.7 |
19 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:berberine bridge enzyme-like protein;
PDBTitle: structure of bbe-like #28 from arabidopsis thaliana
|
|
|
|
| 15 | c3fwaA_
|
|
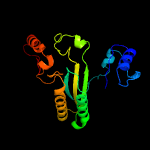 |
99.7 |
15 |
PDB header:flavoprotein
Chain: A: PDB Molecule:reticuline oxidase;
PDBTitle: structure of berberine bridge enzyme, c166a variant in complex with2 (s)-reticuline
|
|
|
|
| 16 | c5l6fA_
|
|
 |
99.7 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:fad linked oxidase-like protein;
PDBTitle: xylooligosaccharide oxidase from myceliophthora thermophila c1 in2 complex with xylobiose
|
|
|
|
| 17 | c1zr6A_
|
|
 |
99.7 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glucooligosaccharide oxidase;
PDBTitle: the crystal structure of an acremonium strictum glucooligosaccharide2 oxidase reveals a novel flavinylation
|
|
|
|
| 18 | c3rjaA_
|
|
 |
99.6 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:carbohydrate oxidase;
PDBTitle: crystal structure of carbohydrate oxidase from microdochium nivale in2 complex with substrate analogue
|
|
|
|
| 19 | c4ml8C_
|
|
 |
99.5 |
11 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:cytokinin oxidase 2;
PDBTitle: structure of maize cytokinin oxidase/dehydrogenase 2 (zmcko2)
|
|
|
|
| 20 | c2bvfA_
|
|
 |
99.5 |
12 |
PDB header:oxidase
Chain: A: PDB Molecule:6-hydroxy-d-nicotine oxidase;
PDBTitle: crystal structure of 6-hydoxy-d-nicotine oxidase from2 arthrobacter nicotinovorans. crystal form 3 (p1)
|
|
|
|
| 21 | c4oalB_ |
|
not modelled |
99.4 |
12 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:cytokinin dehydrogenase 4;
PDBTitle: crystal structure of maize cytokinin oxidase/dehydrogenase 4 (zmcko4)2 in complex with phenylurea inhibitor cppu in alternative spacegroup
|
|
|
| 22 | c6c80B_ |
|
not modelled |
99.4 |
13 |
PDB header:immune system
Chain: B: PDB Molecule:cytokinin oxidase luckx1.1;
PDBTitle: crystal structure of a flax cytokinin oxidase
|
|
|
| 23 | c3bw7A_ |
|
not modelled |
99.4 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:cytokinin dehydrogenase 1;
PDBTitle: maize cytokinin oxidase/dehydrogenase complexed with the allenic2 cytokinin analog ha-1
|
|
|
| 24 | c2exrA_ |
|
not modelled |
99.3 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:cytokinin dehydrogenase 7;
PDBTitle: x-ray structure of cytokinin oxidase/dehydrogenase (ckx) from2 arabidopsis thaliana at5g21482
|
|
|
| 25 | c1wveB_ |
|
not modelled |
99.2 |
13 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:4-cresol dehydrogenase [hydroxylating]
PDBTitle: p-cresol methylhydroxylase: alteration of the structure of2 the flavoprotein subunit upon its binding to the3 cytochrome subunit
|
|
|
| 26 | c3pm9A_ |
|
not modelled |
99.1 |
10 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative oxidoreductase;
PDBTitle: crystal structure of a putative dehydrogenase (rpa1076) from2 rhodopseudomonas palustris cga009 at 2.57 a resolution
|
|
|
| 27 | c4fdoA_ |
|
not modelled |
99.0 |
19 |
PDB header:oxidoreductase/oxidoreductase inhibitor
Chain: A: PDB Molecule:oxidoreductase dpre1;
PDBTitle: mycobacterium tuberculosis dpre1 in complex with ct319
|
|
|
| 28 | c2vfvA_ |
|
not modelled |
99.0 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:xylitol oxidase;
PDBTitle: alditol oxidase from streptomyces coelicolor a3(2): complex2 with sulphite
|
|
|
| 29 | c4bc9C_ |
|
not modelled |
98.9 |
8 |
PDB header:transferase
Chain: C: PDB Molecule:alkyldihydroxyacetonephosphate synthase, peroxisomal;
PDBTitle: mammalian alkyldihydroxyacetonephosphate synthase: wild-type, adduct2 with cyanoethyl
|
|
|
| 30 | c5fxpA_ |
|
not modelled |
98.8 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:eugenol oxidase;
PDBTitle: crystal structure of eugenol oxidase in complex with2 vanillin
|
|
|
| 31 | c3js8A_ |
|
not modelled |
98.8 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:cholesterol oxidase;
PDBTitle: solvent-stable cholesterol oxidase
|
|
|
| 32 | c1i19B_ |
|
not modelled |
98.5 |
11 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:cholesterol oxidase;
PDBTitle: crystal structure of cholesterol oxidase from b.sterolicum
|
|
|
| 33 | c1ahuB_ |
|
not modelled |
98.2 |
15 |
PDB header:flavoenzyme
Chain: B: PDB Molecule:vanillyl-alcohol oxidase;
PDBTitle: structure of the octameric flavoenzyme vanillyl-alcohol2 oxidase in complex with p-cresol
|
|
|
| 34 | d1w1oa1 |
|
not modelled |
96.5 |
18 |
Fold:Ferredoxin-like
Superfamily:FAD-linked oxidases, C-terminal domain
Family:Cytokinin dehydrogenase 1 |
|
|
| 35 | d1wvfa1 |
|
not modelled |
95.9 |
12 |
Fold:Ferredoxin-like
Superfamily:FAD-linked oxidases, C-terminal domain
Family:Vanillyl-alcohol oxidase-like |
|
|
| 36 | c2uuvC_ |
|
not modelled |
95.7 |
8 |
PDB header:transferase
Chain: C: PDB Molecule:alkyldihydroxyacetonephosphate synthase;
PDBTitle: alkyldihydroxyacetonephosphate synthase in p1
|
|
|
| 37 | d1e8ga1 |
|
not modelled |
94.6 |
13 |
Fold:Ferredoxin-like
Superfamily:FAD-linked oxidases, C-terminal domain
Family:Vanillyl-alcohol oxidase-like |
|
|
| 38 | c1f0xA_ |
|
not modelled |
81.4 |
26 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:d-lactate dehydrogenase;
PDBTitle: crystal structure of d-lactate dehydrogenase, a peripheral membrane2 respiratory enzyme.
|
|
|
| 39 | d1f0xa1 |
|
not modelled |
73.6 |
36 |
Fold:Ferredoxin-like
Superfamily:FAD-linked oxidases, C-terminal domain
Family:D-lactate dehydrogenase |
|
|
| 40 | d2i0ka1 |
|
not modelled |
68.3 |
27 |
Fold:Ferredoxin-like
Superfamily:FAD-linked oxidases, C-terminal domain
Family:Cholesterol oxidase |
|
|
| 41 | d1kx9b_ |
|
not modelled |
38.9 |
29 |
Fold:alpha-alpha superhelix
Superfamily:Chemosensory protein Csp2
Family:Chemosensory protein Csp2 |
|
|
| 42 | c2gvsA_ |
|
not modelled |
36.3 |
36 |
PDB header:lipid binding protein
Chain: A: PDB Molecule:chemosensory protein csp-sg4;
PDBTitle: nmr solution structure of cspsg4
|
|
|
| 43 | c5cwsE_ |
|
not modelled |
34.3 |
14 |
PDB header:protein transport
Chain: E: PDB Molecule:nucleoporin nup57;
PDBTitle: crystal structure of the intact chaetomium thermophilum nsp1-nup49-2 nup57 channel nucleoporin heterotrimer bound to its nic96 nuclear3 pore complex attachment site
|
|
|
| 44 | d1n8va_ |
|
not modelled |
32.5 |
29 |
Fold:alpha-alpha superhelix
Superfamily:Chemosensory protein Csp2
Family:Chemosensory protein Csp2 |
|
|
| 45 | c4yh5B_ |
|
not modelled |
31.6 |
17 |
PDB header:transferase
Chain: B: PDB Molecule:levoglucosan kinase;
PDBTitle: lipomyces starkeyi levoglucosan kinase bound to adp and manganese
|
|
|
| 46 | c4guzA_ |
|
not modelled |
30.1 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:probable arylamine n-acetyl transferase;
PDBTitle: structure of the arylamine n-acetyltransferase from mycobacterium2 abscessus
|
|
|
| 47 | d1w4ta1 |
|
not modelled |
26.1 |
32 |
Fold:Cysteine proteinases
Superfamily:Cysteine proteinases
Family:Arylamine N-acetyltransferase |
|
|
| 48 | c3lnbA_ |
|
not modelled |
25.4 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:n-acetyltransferase family protein;
PDBTitle: crystal structure analysis of arylamine n-acetyltransferase c from2 bacillus anthracis
|
|
|
| 49 | d1w5ra1 |
|
not modelled |
24.1 |
20 |
Fold:Cysteine proteinases
Superfamily:Cysteine proteinases
Family:Arylamine N-acetyltransferase |
|
|
| 50 | c2vfbA_ |
|
not modelled |
24.1 |
23 |
PDB header:transferase
Chain: A: PDB Molecule:arylamine n-acetyltransferase;
PDBTitle: the structure of mycobacterium marinum arylamine n-2 acetyltransferase
|
|
|
| 51 | d1e2ta_ |
|
not modelled |
24.0 |
30 |
Fold:Cysteine proteinases
Superfamily:Cysteine proteinases
Family:Arylamine N-acetyltransferase |
|
|
| 52 | d1g7sa2 |
|
not modelled |
23.7 |
25 |
Fold:Reductase/isomerase/elongation factor common domain
Superfamily:Translation proteins
Family:Elongation factors |
|
|
| 53 | d2bsza1 |
|
not modelled |
22.4 |
29 |
Fold:Cysteine proteinases
Superfamily:Cysteine proteinases
Family:Arylamine N-acetyltransferase |
|
|
| 54 | c4dmoB_ |
|
not modelled |
21.6 |
20 |
PDB header:transferase
Chain: B: PDB Molecule:n-hydroxyarylamine o-acetyltransferase;
PDBTitle: crystal structure of the (baccr)nat3 arylamine n-acetyltransferase2 from bacillus cereus reveals a unique cys-his-glu catalytic triad
|
|
|
| 55 | c2pfrB_ |
|
not modelled |
21.4 |
20 |
PDB header:transferase
Chain: B: PDB Molecule:arylamine n-acetyltransferase 2;
PDBTitle: human n-acetyltransferase 2
|
|
|
| 56 | c2dt7A_ |
|
not modelled |
19.9 |
18 |
PDB header:rna binding protein
Chain: A: PDB Molecule:splicing factor 3a subunit 3;
PDBTitle: solution structure of the second surp domain of human2 splicing factor sf3a120 in complex with a fragment of3 human splicing factor sf3a60
|
|
|
| 57 | c3cwyA_ |
|
not modelled |
19.3 |
26 |
PDB header:unknown function
Chain: A: PDB Molecule:protein cagd;
PDBTitle: structure of cagd from h. pylori pathogenicity island crystallized in2 the presence of cu(ii) ions
|
|
|
| 58 | c3d9wA_ |
|
not modelled |
18.2 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:putative acetyltransferase;
PDBTitle: crystal structure analysis of nocardia farcinica arylamine n-2 acetyltransferase
|
|
|
| 59 | d1e89a_ |
|
not modelled |
16.6 |
25 |
Fold:alpha/beta-Hydrolases
Superfamily:alpha/beta-Hydrolases
Family:Hydroxynitrile lyase-like |
|
|
| 60 | c2hcuA_ |
|
not modelled |
16.4 |
33 |
PDB header:lyase
Chain: A: PDB Molecule:3-isopropylmalate dehydratase small subunit;
PDBTitle: crystal structure of smu.1381 (or leud) from streptococcus mutans
|
|
|
| 61 | c3h5jA_ |
|
not modelled |
16.3 |
36 |
PDB header:lyase
Chain: A: PDB Molecule:3-isopropylmalate dehydratase small subunit;
PDBTitle: leud_1-168 small subunit of isopropylmalate isomerase (rv2987c) from2 mycobacterium tuberculosis
|
|
|
| 62 | c3q3wB_ |
|
not modelled |
15.2 |
30 |
PDB header:transferase
Chain: B: PDB Molecule:3-isopropylmalate dehydratase small subunit;
PDBTitle: isopropylmalate isomerase small subunit from campylobacter jejuni.
|
|
|
| 63 | c5a4aA_ |
|
not modelled |
14.7 |
20 |
PDB header:rna binding protein
Chain: A: PDB Molecule:maternal effect protein oskar;
PDBTitle: crystal structure of the osk domain of drosophila oskar
|
|
|
| 64 | c3wwoA_ |
|
not modelled |
14.0 |
21 |
PDB header:lyase
Chain: A: PDB Molecule:(s)-hydroxynitrile lyase;
PDBTitle: s-selective hydroxynitrile lyase from baliospermum montanum (apo1)
|
|
|
| 65 | c5g1mA_ |
|
not modelled |
13.9 |
26 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-hexosaminidase;
PDBTitle: crystal structure of nagz from pseudomonas aeruginosa
|
|
|
| 66 | c3dqzB_ |
|
not modelled |
13.4 |
25 |
PDB header:lyase
Chain: B: PDB Molecule:alpha-hydroxynitrile lyase-like protein;
PDBTitle: structure of the hydroxynitrile lyase from arabidopsis thaliana
|
|
|
| 67 | c6nvoA_ |
|
not modelled |
11.9 |
26 |
PDB header:dna binding protein
Chain: A: PDB Molecule:nuclease mpe;
PDBTitle: crystal structure of pseudomonas putida nuclease mpe
|
|
|
| 68 | c2luyA_ |
|
not modelled |
11.7 |
19 |
PDB header:dna binding protein
Chain: A: PDB Molecule:meiotic chromosome segregation protein p8b7.28c;
PDBTitle: solution structure of the tandem zinc finger domain of fission yeast2 stc1
|
|
|
| 69 | c6fsiA_ |
|
not modelled |
11.7 |
11 |
PDB header:electron transport
Chain: A: PDB Molecule:flavodoxin;
PDBTitle: crystal structure of semiquinone flavodoxin 1 from bacillus cereus2 (1.32 a resolution)
|
|
|
| 70 | c3p2mA_ |
|
not modelled |
11.7 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:possible hydrolase;
PDBTitle: crystal structure of a novel esterase rv0045c from mycobacterium2 tuberculosis
|
|
|
| 71 | c5iobC_ |
|
not modelled |
11.7 |
25 |
PDB header:hydrolase
Chain: C: PDB Molecule:beta-glucosidase-related glycosidases;
PDBTitle: crystal structure of beta-n-acetylglucosaminidase-like protein from2 corynebacterium glutamicum
|
|
|
| 72 | c4gvgB_ |
|
not modelled |
11.1 |
19 |
PDB header:hydrolase
Chain: B: PDB Molecule:beta-hexosaminidase;
PDBTitle: crystal structure of salmonella typhimurium family 3 glycoside2 hydrolase (nagz)
|
|
|
| 73 | d1vfsa2 |
|
not modelled |
10.8 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:PLP-binding barrel
Family:Alanine racemase-like, N-terminal domain |
|
|
| 74 | c3stxB_ |
|
not modelled |
9.8 |
8 |
PDB header:hydrolase
Chain: B: PDB Molecule:methylketone synthase 1;
PDBTitle: crystal structure of tomato methylketone synthase i h243a variant2 complexed with beta-ketoheptanoate
|
|
|
| 75 | c6gwjK_ |
|
not modelled |
9.8 |
9 |
PDB header:rna binding protein
Chain: K: PDB Molecule:probable trna n6-adenosine threonylcarbamoyltransferase;
PDBTitle: protein complex
|
|
|
| 76 | c4em8A_ |
|
not modelled |
9.5 |
67 |
PDB header:isomerase
Chain: A: PDB Molecule:ribose 5-phosphate isomerase b;
PDBTitle: the structure of ribose 5-phosphate isomerase b from anaplasma2 phagocytophilum
|
|
|
| 77 | c3qd5B_ |
|
not modelled |
9.5 |
44 |
PDB header:isomerase
Chain: B: PDB Molecule:putative ribose-5-phosphate isomerase;
PDBTitle: crystal structure of a putative ribose-5-phosphate isomerase from2 coccidioides immitis solved by combined iodide ion sad and mr
|
|
|
| 78 | d1e5da1 |
|
not modelled |
9.4 |
8 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:Flavodoxin-related |
|
|
| 79 | c4lfnD_ |
|
not modelled |
9.4 |
56 |
PDB header:isomerase
Chain: D: PDB Molecule:galactose-6-phosphate isomerase subunit b;
PDBTitle: crystal structure of d-galactose-6-phosphate isomerase in complex with2 d-ribulose
|
|
|
| 80 | d1nn4a_ |
|
not modelled |
9.4 |
33 |
Fold:Ribose/Galactose isomerase RpiB/AlsB
Superfamily:Ribose/Galactose isomerase RpiB/AlsB
Family:Ribose/Galactose isomerase RpiB/AlsB |
|
|
| 81 | c6ahuK_ |
|
not modelled |
9.2 |
38 |
PDB header:hydrolase/rna
Chain: K: PDB Molecule:ribonuclease p protein subunit p21;
PDBTitle: cryo-em structure of human ribonuclease p with mature trna
|
|
|
| 82 | d1g8ka2 |
|
not modelled |
9.2 |
12 |
Fold:Formate dehydrogenase/DMSO reductase, domains 1-3
Superfamily:Formate dehydrogenase/DMSO reductase, domains 1-3
Family:Formate dehydrogenase/DMSO reductase, domains 1-3 |
|
|
| 83 | d1dmra2 |
|
not modelled |
9.0 |
15 |
Fold:Formate dehydrogenase/DMSO reductase, domains 1-3
Superfamily:Formate dehydrogenase/DMSO reductase, domains 1-3
Family:Formate dehydrogenase/DMSO reductase, domains 1-3 |
|
|
| 84 | c4uuzC_ |
|
not modelled |
9.0 |
29 |
PDB header:dna binding protein
Chain: C: PDB Molecule:dna replication licensing factor mcm2;
PDBTitle: mcm2-histone complex
|
|
|
| 85 | c6mu0A_ |
|
not modelled |
8.9 |
22 |
PDB header:isomerase
Chain: A: PDB Molecule:probable ribose-5-phosphate isomerase b;
PDBTitle: crystal structure of ribose-5-phosphate isomerase b from mycoplasma2 genitalium with bound ribulose-5-phosphate
|
|
|
| 86 | c1jjoB_ |
|
not modelled |
8.8 |
19 |
PDB header:signaling protein
Chain: B: PDB Molecule:neuroserpin;
PDBTitle: crystal structure of mouse neuroserpin (cleaved form)
|
|
|
| 87 | c4lfmA_ |
|
not modelled |
8.6 |
11 |
PDB header:isomerase
Chain: A: PDB Molecule:galactose-6-phosphate isomerase subunit a;
PDBTitle: crystal structure of d-galactose-6-phosphate isomerase in complex with2 d-psicose
|
|
|
| 88 | d3c70a1 |
|
not modelled |
8.4 |
24 |
Fold:alpha/beta-Hydrolases
Superfamily:alpha/beta-Hydrolases
Family:Hydroxynitrile lyase-like |
|
|
| 89 | c2pkpA_ |
|
not modelled |
8.2 |
22 |
PDB header:lyase
Chain: A: PDB Molecule:homoaconitase small subunit;
PDBTitle: crystal structure of 3-isopropylmalate dehydratase (leud)2 from methhanocaldococcus jannaschii dsm2661 (mj1271)
|
|
|
| 90 | c3m1pA_ |
|
not modelled |
8.2 |
56 |
PDB header:isomerase
Chain: A: PDB Molecule:ribose 5-phosphate isomerase;
PDBTitle: structure of ribose 5-phosphate isomerase type b from trypanosoma2 cruzi, soaked with allose-6-phosphate
|
|
|
| 91 | c3k7pA_ |
|
not modelled |
8.2 |
56 |
PDB header:isomerase
Chain: A: PDB Molecule:ribose 5-phosphate isomerase;
PDBTitle: structure of mutant of ribose 5-phosphate isomerase type b from2 trypanosoma cruzi.
|
|
|
| 92 | c6ah3K_ |
|
not modelled |
8.1 |
10 |
PDB header:hydrolase/rna
Chain: K: PDB Molecule:ribonuclease p protein subunit rpr2;
PDBTitle: cryo-em structure of yeast ribonuclease p with pre-trna substrate
|
|
|
| 93 | c6fxwA_ |
|
not modelled |
8.1 |
67 |
PDB header:isomerase
Chain: A: PDB Molecule:putative ribose 5-phosphate isomerase;
PDBTitle: structure of leishmania infantum type b ribose 5-phosphate isomerase
|
|
|
| 94 | c1g8jC_ |
|
not modelled |
8.1 |
10 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:arsenite oxidase;
PDBTitle: crystal structure analysis of arsenite oxidase from2 alcaligenes faecalis
|
|
|
| 95 | c3he8A_ |
|
not modelled |
8.0 |
56 |
PDB header:isomerase
Chain: A: PDB Molecule:ribose-5-phosphate isomerase;
PDBTitle: structural study of clostridium thermocellum ribose-5-phosphate2 isomerase b
|
|
|
| 96 | c3vbaE_ |
|
not modelled |
8.0 |
31 |
PDB header:lyase
Chain: E: PDB Molecule:isopropylmalate/citramalate isomerase small subunit;
PDBTitle: crystal structure of methanogen 3-isopropylmalate isomerase small2 subunit
|
|
|
| 97 | c3s5pA_ |
|
not modelled |
7.9 |
67 |
PDB header:isomerase
Chain: A: PDB Molecule:ribose 5-phosphate isomerase;
PDBTitle: crystal structure of ribose-5-phosphate isomerase b rpib from giardia2 lamblia
|
|
|
| 98 | c4lidA_ |
|
not modelled |
7.8 |
21 |
PDB header:viral protein
Chain: A: PDB Molecule:a-100;
PDBTitle: a100, a dna binding scaffold from sulfolobus spindle-shape virus 1
|
|
|
| 99 | c3j20F_ |
|
not modelled |
7.8 |
26 |
PDB header:ribosome
Chain: F: PDB Molecule:30s ribosomal protein s5p;
PDBTitle: promiscuous behavior of proteins in archaeal ribosomes revealed by2 cryo-em: implications for evolution of eukaryotic ribosomes (30s3 ribosomal subunit)
|
|
|