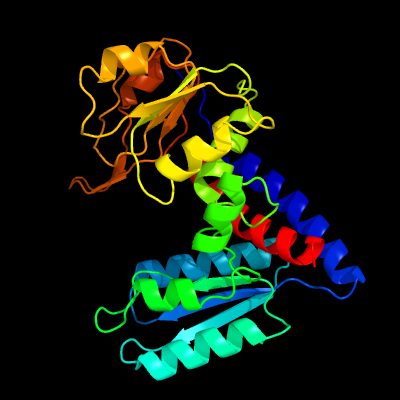
| 1 | c2c2xB_
|
|
 |
100.0 |
100 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:methylenetetrahydrofolate dehydrogenase-
PDBTitle: three dimensional structure of bifunctional methylenetetrahydrofolate2 dehydrogenase-cyclohydrolase from mycobacterium tuberculosis
|
|
|
|
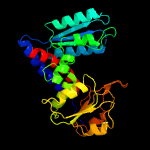
| 2 | c4b4uB_
|
|
 |
100.0 |
45 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:bifunctional protein fold;
PDBTitle: crystal structure of acinetobacter baumannii n5,2 n10-methylenetetrahydrofolate dehydrogenase-cyclohydrolase (fold)3 complexed with nadp cofactor
|
|
|
|
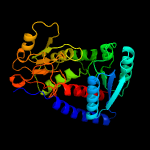
| 3 | c4cjxA_
|
|
 |
100.0 |
42 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:c-1-tetrahydrofolate synthase, cytoplasmic, putative;
PDBTitle: the crystal structure of trypanosoma brucei n5, n10-2 methylenetetrahydrofolate dehydrogenase-cyclohydrolase (fold)3 complexed with nadp cofactor and inhibitor
|
|
|
|
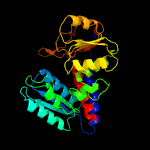
| 4 | c1b0aA_
|
|
 |
100.0 |
43 |
PDB header:oxidoreductase,hydrolase
Chain: A: PDB Molecule:protein (fold bifunctional protein);
PDBTitle: 5,10, methylene-tetrahydropholate2 dehydrogenase/cyclohydrolase from e coli.
|
|
|
|
| 5 | c4a5oB_
|
|
 |
100.0 |
46 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:bifunctional protein fold;
PDBTitle: crystal structure of pseudomonas aeruginosa n5, n10-2 methylenetetrahydrofolate dehydrogenase-cyclohydrolase (fold)
|
|
|
|
| 6 | c3l07B_
|
|
 |
100.0 |
42 |
PDB header:oxidoreductase,hydrolase
Chain: B: PDB Molecule:bifunctional protein fold;
PDBTitle: methylenetetrahydrofolate dehydrogenase/methenyltetrahydrofolate2 cyclohydrolase, putative bifunctional protein fold from francisella3 tularensis.
|
|
|
|
| 7 | c1a4iB_
|
|
 |
100.0 |
38 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:methylenetetrahydrofolate dehydrogenase /
PDBTitle: human tetrahydrofolate dehydrogenase / cyclohydrolase
|
|
|
|
| 8 | c6apeA_
|
|
 |
100.0 |
39 |
PDB header:oxidoreductase, hydrolase
Chain: A: PDB Molecule:bifunctional protein fold;
PDBTitle: crystal structure of bifunctional protein fold from helicobacter2 pylori
|
|
|
|
| 9 | c3p2oA_
|
|
 |
100.0 |
42 |
PDB header:oxidoreductase, hydrolase
Chain: A: PDB Molecule:bifunctional protein fold;
PDBTitle: crystal structure of fold bifunctional protein from campylobacter2 jejuni
|
|
|
|
| 10 | c3p2oB_
|
|
 |
100.0 |
42 |
PDB header:oxidoreductase, hydrolase
Chain: B: PDB Molecule:bifunctional protein fold;
PDBTitle: crystal structure of fold bifunctional protein from campylobacter2 jejuni
|
|
|
|
| 11 | c3nglA_
|
|
 |
100.0 |
36 |
PDB header:oxidoreductase, hydrolase
Chain: A: PDB Molecule:bifunctional protein fold;
PDBTitle: crystal structure of bifunctional 5,10-methylenetetrahydrofolate2 dehydrogenase / cyclohydrolase from thermoplasma acidophilum
|
|
|
|
| 12 | c5zf1A_
|
|
 |
100.0 |
42 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:5,10-methylenetetrahydrofolate dehydrogenase;
PDBTitle: molecular structure of a novel 5,10-methylenetetrahydrofolate2 dehydrogenase from the silkworm, bombyx mori
|
|
|
|
| 13 | c5tc4A_
|
|
 |
100.0 |
42 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:bifunctional methylenetetrahydrofolate
PDBTitle: crystal structure of human mitochondrial methylenetetrahydrofolate2 dehydrogenase-cyclohydrolase (mthfd2) in complex with ly345899 and3 cofactors
|
|
|
|
| 14 | c4a26B_
|
|
 |
100.0 |
47 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative c-1-tetrahydrofolate synthase, cytoplasmic;
PDBTitle: the crystal structure of leishmania major n5,n10-2 methylenetetrahydrofolate dehydrogenase/cyclohydrolase
|
|
|
|
| 15 | c5nhsB_
|
|
 |
100.0 |
46 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:bifunctional protein fold;
PDBTitle: the crystal structure of xanthomonas albilineans n5, n10-2 methylenetetrahydrofolate dehydrogenase-cyclohydrolase (fold)
|
|
|
|
| 16 | c1edzA_
|
|
 |
100.0 |
26 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:5,10-methylenetetrahydrofolate dehydrogenase;
PDBTitle: structure of the nad-dependent 5,10-2 methylenetetrahydrofolate dehydrogenase from saccharomyces3 cerevisiae
|
|
|
|
| 17 | d1b0aa1
|
|
 |
100.0 |
42 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Aminoacid dehydrogenase-like, C-terminal domain |
|
|
|
| 18 | d1a4ia1
|
|
 |
100.0 |
43 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Aminoacid dehydrogenase-like, C-terminal domain |
|
|
|
| 19 | d1edza2
|
|
 |
100.0 |
17 |
Fold:Aminoacid dehydrogenase-like, N-terminal domain
Superfamily:Aminoacid dehydrogenase-like, N-terminal domain
Family:Tetrahydrofolate dehydrogenase/cyclohydrolase |
|
|
|
| 20 | d1edza1
|
|
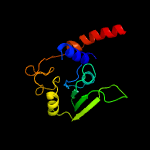 |
100.0 |
30 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Aminoacid dehydrogenase-like, C-terminal domain |
|
|
|
| 21 | d1b0aa2 |
|
not modelled |
100.0 |
45 |
Fold:Aminoacid dehydrogenase-like, N-terminal domain
Superfamily:Aminoacid dehydrogenase-like, N-terminal domain
Family:Tetrahydrofolate dehydrogenase/cyclohydrolase |
|
|
| 22 | d1a4ia2 |
|
not modelled |
100.0 |
30 |
Fold:Aminoacid dehydrogenase-like, N-terminal domain
Superfamily:Aminoacid dehydrogenase-like, N-terminal domain
Family:Tetrahydrofolate dehydrogenase/cyclohydrolase |
|
|
| 23 | c3d4oA_ |
|
not modelled |
99.9 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dipicolinate synthase subunit a;
PDBTitle: crystal structure of dipicolinate synthase subunit a (np_243269.1)2 from bacillus halodurans at 2.10 a resolution
|
|
|
| 24 | c1v8bA_ |
|
not modelled |
99.9 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:adenosylhomocysteinase;
PDBTitle: crystal structure of a hydrolase
|
|
|
| 25 | c2rirA_ |
|
not modelled |
99.9 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dipicolinate synthase, a chain;
PDBTitle: crystal structure of dipicolinate synthase, a chain, from bacillus2 subtilis
|
|
|
| 26 | c3oneA_ |
|
not modelled |
99.5 |
19 |
PDB header:hydrolase/hydrolase substrate
Chain: A: PDB Molecule:adenosylhomocysteinase;
PDBTitle: crystal structure of lupinus luteus s-adenosyl-l-homocysteine2 hydrolase in complex with adenine
|
|
|
| 27 | c3x2fA_ |
|
not modelled |
99.1 |
24 |
PDB header:hydrolase
Chain: A: PDB Molecule:adenosylhomocysteinase;
PDBTitle: a thermophilic s-adenosylhomocysteine hydrolase
|
|
|
| 28 | c6aphA_ |
|
not modelled |
99.1 |
24 |
PDB header:hydrolase
Chain: A: PDB Molecule:adenosylhomocysteinase;
PDBTitle: crystal structure of adenosylhomocysteinase from elizabethkingia2 anophelis nuhp1 in complex with nad and adenosine
|
|
|
| 29 | d1li4a1 |
|
not modelled |
99.0 |
22 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Formate/glycerate dehydrogenases, NAD-domain |
|
|
| 30 | d1v8ba1 |
|
not modelled |
99.0 |
22 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Formate/glycerate dehydrogenases, NAD-domain |
|
|
| 31 | c1d4fD_ |
|
not modelled |
98.9 |
22 |
PDB header:hydrolase
Chain: D: PDB Molecule:s-adenosylhomocysteine hydrolase;
PDBTitle: crystal structure of recombinant rat-liver d244e mutant s-2 adenosylhomocysteine hydrolase
|
|
|
| 32 | c3gvpB_ |
|
not modelled |
98.9 |
27 |
PDB header:hydrolase
Chain: B: PDB Molecule:adenosylhomocysteinase 3;
PDBTitle: human sahh-like domain of human adenosylhomocysteinase 3
|
|
|
| 33 | c3d64A_ |
|
not modelled |
98.8 |
23 |
PDB header:hydrolase
Chain: A: PDB Molecule:adenosylhomocysteinase;
PDBTitle: crystal structure of s-adenosyl-l-homocysteine hydrolase from2 burkholderia pseudomallei
|
|
|
| 34 | c3n58D_ |
|
not modelled |
98.8 |
29 |
PDB header:hydrolase
Chain: D: PDB Molecule:adenosylhomocysteinase;
PDBTitle: crystal structure of s-adenosyl-l-homocysteine hydrolase from brucella2 melitensis in ternary complex with nad and adenosine, orthorhombic3 form
|
|
|
| 35 | c5v96A_ |
|
not modelled |
98.7 |
28 |
PDB header:hydrolase
Chain: A: PDB Molecule:s-adenosyl-l-homocysteine hydrolase;
PDBTitle: crystal structure of s-adenosyl-l-homocysteine hydrolase from2 naegleria fowleri with bound nad and adenosine
|
|
|
| 36 | c1gpjA_ |
|
not modelled |
98.7 |
26 |
PDB header:reductase
Chain: A: PDB Molecule:glutamyl-trna reductase;
PDBTitle: glutamyl-trna reductase from methanopyrus kandleri
|
|
|
| 37 | d1gpja2 |
|
not modelled |
98.5 |
29 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Aminoacid dehydrogenase-like, C-terminal domain |
|
|
| 38 | c3dhyC_ |
|
not modelled |
98.5 |
24 |
PDB header:hydrolase
Chain: C: PDB Molecule:adenosylhomocysteinase;
PDBTitle: crystal structures of mycobacterium tuberculosis s-adenosyl-l-2 homocysteine hydrolase in ternary complex with substrate and3 inhibitors
|
|
|
| 39 | c5hm8C_ |
|
not modelled |
98.5 |
26 |
PDB header:hydrolase
Chain: C: PDB Molecule:adenosylhomocysteinase;
PDBTitle: 2.85 angstrom crystal structure of s-adenosylhomocysteinase from2 cryptosporidium parvum in complex with adenosine and nad.
|
|
|
| 40 | c2dbqA_ |
|
not modelled |
98.5 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glyoxylate reductase;
PDBTitle: crystal structure of glyoxylate reductase (ph0597) from pyrococcus2 horikoshii ot3, complexed with nadp (i41)
|
|
|
| 41 | c4njmA_ |
|
not modelled |
98.3 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:d-3-phosphoglycerate dehydrogenase, putative;
PDBTitle: crystal structure of phosphoglycerate bound 3-phosphoglycerate2 dehydrogenase in entamoeba histolytica
|
|
|
| 42 | c6f3oC_ |
|
not modelled |
98.3 |
26 |
PDB header:hydrolase
Chain: C: PDB Molecule:adenosylhomocysteinase;
PDBTitle: crystal structure of s-adenosyl-l-homocysteine hydrolase from2 pseudomonas aeruginosa complexed with adenine, k+ and zn2+ cations
|
|
|
| 43 | c3oj0A_ |
|
not modelled |
98.3 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glutamyl-trna reductase;
PDBTitle: crystal structure of glutamyl-trna reductase from thermoplasma2 volcanium (nucleotide binding domain)
|
|
|
| 44 | c2eklA_ |
|
not modelled |
98.3 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:d-3-phosphoglycerate dehydrogenase;
PDBTitle: structure of st1218 protein from sulfolobus tokodaii
|
|
|
| 45 | c3fbtB_ |
|
not modelled |
98.2 |
18 |
PDB header:oxidoreductase, lyase
Chain: B: PDB Molecule:chorismate mutase and shikimate 5-dehydrogenase fusion
PDBTitle: crystal structure of a chorismate mutase/shikimate 5-dehydrogenase2 fusion protein from clostridium acetobutylicum
|
|
|
| 46 | c6ih2B_ |
|
not modelled |
98.2 |
20 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:phosphite dehydrogenase;
PDBTitle: crystal structure of phosphite dehydrogenase from ralstonia sp. 4506
|
|
|
| 47 | c3tozA_ |
|
not modelled |
98.2 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:shikimate dehydrogenase;
PDBTitle: 2.2 angstrom crystal structure of shikimate 5-dehydrogenase from2 listeria monocytogenes in complex with nad.
|
|
|
| 48 | c2j6iC_ |
|
not modelled |
98.2 |
19 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:formate dehydrogenase;
PDBTitle: candida boidinii formate dehydrogenase (fdh) c-terminal mutant
|
|
|
| 49 | c4n7rB_ |
|
not modelled |
98.2 |
26 |
PDB header:oxidoreductase/protein binding
Chain: B: PDB Molecule:glutamyl-trna reductase 1, chloroplastic;
PDBTitle: crystal structure of arabidopsis glutamyl-trna reductase in complex2 with its binding protein
|
|
|
| 50 | c3pgjB_ |
|
not modelled |
98.2 |
18 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:shikimate dehydrogenase;
PDBTitle: 2.49 angstrom resolution crystal structure of shikimate 5-2 dehydrogenase (aroe) from vibrio cholerae o1 biovar eltor str. n169613 in complex with shikimate
|
|
|
| 51 | c1xdwA_ |
|
not modelled |
98.2 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nad+-dependent (r)-2-hydroxyglutarate
PDBTitle: nad+-dependent (r)-2-hydroxyglutarate dehydrogenase from2 acidaminococcus fermentans
|
|
|
| 52 | c2o4cB_ |
|
not modelled |
98.2 |
25 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:erythronate-4-phosphate dehydrogenase;
PDBTitle: crystal structure of d-erythronate-4-phosphate dehydrogenase complexed2 with nad
|
|
|
| 53 | c2nloA_ |
|
not modelled |
98.1 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:shikimate dehydrogenase;
PDBTitle: crystal structure of the quinate dehydrogenase from corynebacterium2 glutamicum
|
|
|
| 54 | c3n7uD_ |
|
not modelled |
98.1 |
15 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:formate dehydrogenase;
PDBTitle: nad-dependent formate dehydrogenase from higher-plant arabidopsis2 thaliana in complex with nad and azide
|
|
|
| 55 | c1p74B_ |
|
not modelled |
98.1 |
22 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:shikimate 5-dehydrogenase;
PDBTitle: crystal structure of shikimate dehydrogenase (aroe) from haemophilus2 influenzae
|
|
|
| 56 | c2hk8B_ |
|
not modelled |
98.1 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:shikimate dehydrogenase;
PDBTitle: crystal structure of shikimate dehydrogenase from aquifex2 aeolicus at 2.35 angstrom resolution
|
|
|
| 57 | d1l7da1 |
|
not modelled |
98.1 |
24 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Formate/glycerate dehydrogenases, NAD-domain |
|
|
| 58 | c3tumA_ |
|
not modelled |
98.1 |
29 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:shikimate dehydrogenase family protein;
PDBTitle: 2.15 angstrom resolution crystal structure of a shikimate2 dehydrogenase family protein from pseudomonas putida kt2440 in3 complex with nad+
|
|
|
| 59 | c2eggA_ |
|
not modelled |
98.1 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:shikimate 5-dehydrogenase;
PDBTitle: crystal structure of shikimate 5-dehydrogenase (aroe) from geobacillus2 kaustophilus
|
|
|
| 60 | c3o8qB_ |
|
not modelled |
98.1 |
17 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:shikimate 5-dehydrogenase i alpha;
PDBTitle: 1.45 angstrom resolution crystal structure of shikimate 5-2 dehydrogenase (aroe) from vibrio cholerae
|
|
|
| 61 | c3oetF_ |
|
not modelled |
98.0 |
25 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:erythronate-4-phosphate dehydrogenase;
PDBTitle: d-erythronate-4-phosphate dehydrogenase complexed with nad
|
|
|
| 62 | c2cukC_ |
|
not modelled |
98.0 |
18 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:glycerate dehydrogenase/glyoxylate reductase;
PDBTitle: crystal structure of tt0316 protein from thermus thermophilus hb8
|
|
|
| 63 | c3donA_ |
|
not modelled |
97.9 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:shikimate dehydrogenase;
PDBTitle: crystal structure of shikimate dehydrogenase from staphylococcus2 epidermidis
|
|
|
| 64 | c4mp6A_ |
|
not modelled |
97.9 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative ornithine cyclodeaminase;
PDBTitle: staphyloferrin b precursor biosynthetic enzyme sbnb bound to citrate2 and nad+
|
|
|
| 65 | c2gcgB_ |
|
not modelled |
97.9 |
20 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:glyoxylate reductase/hydroxypyruvate reductase;
PDBTitle: ternary crystal structure of human glyoxylate2 reductase/hydroxypyruvate reductase
|
|
|
| 66 | c1nvtA_ |
|
not modelled |
97.9 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:shikimate 5'-dehydrogenase;
PDBTitle: crystal structure of shikimate dehydrogenase (aroe or2 mj1084) in complex with nadp+
|
|
|
| 67 | c5j23D_ |
|
not modelled |
97.9 |
21 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:2-hydroxyacid dehydrogenase;
PDBTitle: crystal structure of nadph-dependent glyoxylate/hydroxypyruvate2 reductase smc04462 (smghrb) from sinorhizobium meliloti in complex3 with 2'-phospho-adp-ribose
|
|
|
| 68 | c1gdhA_ |
|
not modelled |
97.9 |
13 |
PDB header:oxidoreductase(choh (d)-nad(p)+ (a))
Chain: A: PDB Molecule:d-glycerate dehydrogenase;
PDBTitle: crystal structure of a nad-dependent d-glycerate2 dehydrogenase at 2.4 angstroms resolution
|
|
|
| 69 | c1nytC_ |
|
not modelled |
97.9 |
19 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:shikimate 5-dehydrogenase;
PDBTitle: shikimate dehydrogenase aroe complexed with nadp+
|
|
|
| 70 | c4n18A_ |
|
not modelled |
97.9 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:d-isomer specific 2-hydroxyacid dehydrogenase family
PDBTitle: crystal structure of d-isomer specific 2-hydroxyacid dehydrogenase2 family protein from klebsiella pneumoniae 342
|
|
|
| 71 | c3p2yA_ |
|
not modelled |
97.8 |
29 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:alanine dehydrogenase/pyridine nucleotide transhydrogenase;
PDBTitle: crystal structure of alanine dehydrogenase/pyridine nucleotide2 transhydrogenase from mycobacterium smegmatis
|
|
|
| 72 | c2eezG_ |
|
not modelled |
97.8 |
29 |
PDB header:oxidoreductase
Chain: G: PDB Molecule:alanine dehydrogenase;
PDBTitle: crystal structure of alanine dehydrogenase from themus thermophilus
|
|
|
| 73 | d1pjca1 |
|
not modelled |
97.8 |
29 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Formate/glycerate dehydrogenases, NAD-domain |
|
|
| 74 | c4e5kC_ |
|
not modelled |
97.8 |
23 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:phosphite dehydrogenase (thermostable variant);
PDBTitle: thermostable phosphite dehydrogenase in complex with nad and sulfite
|
|
|
| 75 | c5dt9A_ |
|
not modelled |
97.8 |
25 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:erythronate-4-phosphate dehydrogenase;
PDBTitle: crystal structure of a putative d-erythronate-4-phosphate2 dehydrogenase from vibrio cholerae
|
|
|
| 76 | d1ygya1 |
|
not modelled |
97.8 |
22 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Formate/glycerate dehydrogenases, NAD-domain |
|
|
| 77 | c1j4aA_ |
|
not modelled |
97.8 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:d-lactate dehydrogenase;
PDBTitle: insights into domain closure, substrate specificity and2 catalysis of d-lactate dehydrogenase from lactobacillus3 bulgaricus
|
|
|
| 78 | c5tx7A_ |
|
not modelled |
97.8 |
27 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:d-isomer specific 2-hydroxyacid dehydrogenase family
PDBTitle: crystal structure of d-isomer specific 2-hydroxyacid dehydrogenase2 from desulfovibrio vulgaris
|
|
|
| 79 | c3bazA_ |
|
not modelled |
97.8 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:hydroxyphenylpyruvate reductase;
PDBTitle: structure of hydroxyphenylpyruvate reductase from coleus blumei in2 complex with nadp+
|
|
|
| 80 | c1wwkA_ |
|
not modelled |
97.7 |
23 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:phosphoglycerate dehydrogenase;
PDBTitle: crystal structure of phosphoglycerate dehydrogenase from pyrococcus2 horikoshii ot3
|
|
|
| 81 | d1gdha1 |
|
not modelled |
97.7 |
13 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Formate/glycerate dehydrogenases, NAD-domain |
|
|
| 82 | d1vi2a1 |
|
not modelled |
97.7 |
17 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Aminoacid dehydrogenase-like, C-terminal domain |
|
|
| 83 | c4dioB_ |
|
not modelled |
97.7 |
23 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:nad(p) transhydrogenase subunit alpha part 1;
PDBTitle: the crystal structure of transhydrogenase from sinorhizobium meliloti
|
|
|
| 84 | d1mx3a1 |
|
not modelled |
97.7 |
27 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Formate/glycerate dehydrogenases, NAD-domain |
|
|
| 85 | c2g76A_ |
|
not modelled |
97.7 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:d-3-phosphoglycerate dehydrogenase;
PDBTitle: crystal structure of human 3-phosphoglycerate dehydrogenase
|
|
|
| 86 | c3pwzA_ |
|
not modelled |
97.6 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:shikimate dehydrogenase 3;
PDBTitle: crystal structure of an ael1 enzyme from pseudomonas putida
|
|
|
| 87 | d2naca1 |
|
not modelled |
97.6 |
12 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Formate/glycerate dehydrogenases, NAD-domain |
|
|
| 88 | c3wnvA_ |
|
not modelled |
97.6 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glyoxylate reductase;
PDBTitle: crystal structure of a glyoxylate reductase from paecilomyes2 thermophila
|
|
|
| 89 | c4xa8A_ |
|
not modelled |
97.6 |
27 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:d-isomer specific 2-hydroxyacid dehydrogenase nad-binding;
PDBTitle: crystal structure of d-isomer specific 2-hydroxyacid dehydrogenase2 from xanthobacter autotrophicus py2
|
|
|
| 90 | c5mh5A_ |
|
not modelled |
97.6 |
23 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:d-2-hydroxyacid dehydrogenase;
PDBTitle: d-2-hydroxyacid dehydrogenases (d2-hdh) from haloferax mediterranei in2 complex with 2-keto-hexanoic acid and nadp+ (1.4 a resolution)
|
|
|
| 91 | c3evtA_ |
|
not modelled |
97.6 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:phosphoglycerate dehydrogenase;
PDBTitle: crystal structure of phosphoglycerate dehydrogenase from lactobacillus2 plantarum
|
|
|
| 92 | c2omeA_ |
|
not modelled |
97.6 |
25 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:c-terminal-binding protein 2;
PDBTitle: crystal structure of human ctbp2 dehydrogenase complexed with nad(h)
|
|
|
| 93 | d1nvta1 |
|
not modelled |
97.6 |
17 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Aminoacid dehydrogenase-like, C-terminal domain |
|
|
| 94 | d1qp8a1 |
|
not modelled |
97.6 |
29 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Formate/glycerate dehydrogenases, NAD-domain |
|
|
| 95 | d1j4aa1 |
|
not modelled |
97.6 |
19 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Formate/glycerate dehydrogenases, NAD-domain |
|
|
| 96 | c2ev9B_ |
|
not modelled |
97.6 |
22 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:shikimate 5-dehydrogenase;
PDBTitle: crystal structure of shikimate 5-dehydrogenase (aroe) from thermus2 thermophilus hb8 in complex with nadp(h) and shikimate
|
|
|
| 97 | c3wwzB_ |
|
not modelled |
97.6 |
30 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:d-lactate dehydrogenase (fermentative);
PDBTitle: the crystal structure of d-lactate dehydrogenase from pseudomonas2 aeruginosa
|
|
|
| 98 | c3fn4A_ |
|
not modelled |
97.5 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nad-dependent formate dehydrogenase;
PDBTitle: apo-form of nad-dependent formate dehydrogenase from bacterium2 moraxella sp.c-1 in closed conformation
|
|
|
| 99 | c4zqbB_ |
|
not modelled |
97.5 |
22 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:nadp-dependent dehydrogenase;
PDBTitle: crystal structure of nadp-dependent dehydrogenase from2 rhodobactersphaeroides in complex with nadp and sulfate
|
|
|
| 100 | c4weqA_ |
|
not modelled |
97.5 |
23 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nad-dependent dehydrogenase;
PDBTitle: crystal structure of nadph-dependent glyoxylate/hydroxypyruvate2 reductase smc02828 (smghra) from sinorhizobium meliloti in complex3 with nadp and sulfate
|
|
|
| 101 | c4g2nA_ |
|
not modelled |
97.5 |
23 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:d-isomer specific 2-hydroxyacid dehydrogenase, nad-binding;
PDBTitle: crystal structure of putative d-isomer specific 2-hydroxyacid2 dehydrogenase, nad-binding from polaromonas sp. js6 66
|
|
|
| 102 | c3hg7A_ |
|
not modelled |
97.5 |
27 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:d-isomer specific 2-hydroxyacid dehydrogenase family
PDBTitle: crystal structure of d-isomer specific 2-hydroxyacid dehydrogenase2 family protein from aeromonas salmonicida subsp. salmonicida a449
|
|
|
| 103 | d1dxya1 |
|
not modelled |
97.5 |
23 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Formate/glycerate dehydrogenases, NAD-domain |
|
|
| 104 | c3gvxA_ |
|
not modelled |
97.5 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glycerate dehydrogenase related protein;
PDBTitle: crystal structure of glycerate dehydrogenase related2 protein from thermoplasma acidophilum
|
|
|
| 105 | c3kboB_ |
|
not modelled |
97.5 |
17 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:glyoxylate/hydroxypyruvate reductase a;
PDBTitle: 2.14 angstrom crystal structure of putative oxidoreductase (ycdw) from2 salmonella typhimurium in complex with nadp
|
|
|
| 106 | c4a7pA_ |
|
not modelled |
97.5 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:udp-glucose dehydrogenase;
PDBTitle: se-met derivatized ugdg, udp-glucose dehydrogenase from sphingomonas2 elodea
|
|
|
| 107 | c2pi1C_ |
|
not modelled |
97.5 |
18 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:d-lactate dehydrogenase;
PDBTitle: crystal structure of d-lactate dehydrogenase from aquifex2 aeolicus complexed with nad and lactic acid
|
|
|
| 108 | c4lswA_ |
|
not modelled |
97.5 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:d-2-hydroxyacid dehydrogensase protein;
PDBTitle: crystallization and structural analysis of 2-hydroxyacid dehydrogenase2 from ketogulonicigenium vulgare y25
|
|
|
| 109 | c2bruB_ |
|
not modelled |
97.4 |
26 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:nad(p) transhydrogenase subunit alpha;
PDBTitle: complex of the domain i and domain iii of escherichia coli2 transhydrogenase
|
|
|
| 110 | c1pjcA_ |
|
not modelled |
97.4 |
26 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:protein (l-alanine dehydrogenase);
PDBTitle: l-alanine dehydrogenase complexed with nad
|
|
|
| 111 | d1nyta1 |
|
not modelled |
97.4 |
20 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Aminoacid dehydrogenase-like, C-terminal domain |
|
|
| 112 | c3k5pA_ |
|
not modelled |
97.4 |
25 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:d-3-phosphoglycerate dehydrogenase;
PDBTitle: crystal structure of amino acid-binding act: d-isomer specific 2-2 hydroxyacid dehydrogenase catalytic domain from brucella melitensis
|
|
|
| 113 | c3wwyA_ |
|
not modelled |
97.4 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:d-lactate dehydrogenase;
PDBTitle: the crystal structure of d-lactate dehydrogenase from fusobacterium2 nucleatum subsp. nucleatum
|
|
|
| 114 | c4omuA_ |
|
not modelled |
97.4 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:shikimate dehydrogenase;
PDBTitle: crystal structure of shikimate dehydrogenase (aroe) from pseudomonas2 putida
|
|
|
| 115 | c2nacA_ |
|
not modelled |
97.4 |
14 |
PDB header:oxidoreductase(aldehyde(d),nad+(a))
Chain: A: PDB Molecule:nad-dependent formate dehydrogenase;
PDBTitle: high resolution structures of holo and apo formate dehydrogenase
|
|
|
| 116 | c4zgsE_ |
|
not modelled |
97.4 |
21 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:putative d-lactate dehydrogenase;
PDBTitle: identification of the pyruvate reductase of chlamydomonas reinhardtii
|
|
|
| 117 | c4cukA_ |
|
not modelled |
97.4 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:d-lactate dehydrogenase;
PDBTitle: structure of salmonella d-lactate dehydrogenase in complex2 with nadh
|
|
|
| 118 | c4s1vD_ |
|
not modelled |
97.3 |
12 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:d-3-phosphoglycerate dehydrogenase-related protein;
PDBTitle: crystal structure of phosphoglycerate oxidoreductase from vibrio2 cholerae o395
|
|
|
| 119 | c2yq4C_ |
|
not modelled |
97.3 |
19 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:d-isomer specific 2-hydroxyacid dehydrogenase;
PDBTitle: crystal structure of d-isomer specific 2-hydroxyacid dehydrogenase2 from lactobacillus delbrueckii ssp. bulgaricus
|
|
|
| 120 | c1luaA_ |
|
not modelled |
97.3 |
26 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:methylene tetrahydromethanopterin dehydrogenase;
PDBTitle: structure of methylene-tetrahydromethanopterin dehydrogenase from2 methylobacterium extorquens am1 complexed with nadp
|
|
|