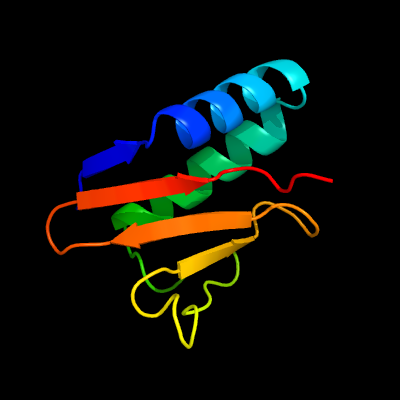
| 1 | c3oeiH_
|
|
 |
100.0 |
100 |
PDB header:toxin, protein binding
Chain: H: PDB Molecule:relk (toxin rv3358);
PDBTitle: crystal structure of mycobacterium tuberculosis reljk (rv3357-rv3358-2 relbe3)
|
|
|
|
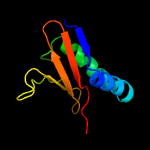
| 2 | d2a6sa1
|
|
 |
100.0 |
45 |
Fold:RelE-like
Superfamily:RelE-like
Family:YoeB/Txe-like |
|
|
|
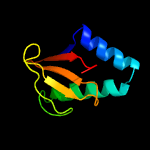
| 3 | c4q2uH_
|
|
 |
99.9 |
18 |
PDB header:toxin/toxin repressor
Chain: H: PDB Molecule:mrna interferase yafq;
PDBTitle: crystal structure of the e. coli dinj-yafq toxin-antitoxin complex
|
|
|
|
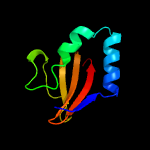
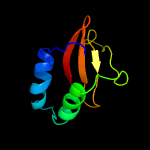
| 4 | d1z8ma1
|
|
 |
99.9 |
20 |
Fold:RelE-like
Superfamily:RelE-like
Family:RelE-like |
|
|
|
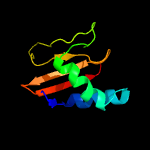
| 5 | c2otrA_
|
|
 |
99.9 |
14 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein hp0892;
PDBTitle: solution structure of conserved hypothetical protein hp0892 from2 helicobacter pylori
|
|
|
|
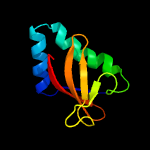
| 6 | c2kheA_
|
|
 |
99.4 |
33 |
PDB header:hydrolase
Chain: A: PDB Molecule:toxin-like protein;
PDBTitle: solution structure of the bacterial toxin rele from thermus2 thermophilus hb8
|
|
|
|
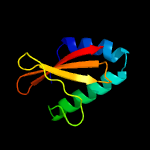
| 7 | c3bpqD_
|
|
 |
99.3 |
17 |
PDB header:toxin
Chain: D: PDB Molecule:toxin rele3;
PDBTitle: crystal structure of relb-rele antitoxin-toxin complex from2 methanococcus jannaschii
|
|
|
|
| 8 | d1wmia1
|
|
 |
99.2 |
15 |
Fold:RelE-like
Superfamily:RelE-like
Family:RelE-like |
|
|
|
| 9 | c3g5oC_
|
|
 |
99.2 |
27 |
PDB header:toxin/antitoxin
Chain: C: PDB Molecule:uncharacterized protein rv2866;
PDBTitle: the crystal structure of the toxin-antitoxin complex relbe2 (rv2865-2 2866) from mycobacterium tuberculosis
|
|
|
|
| 10 | c3kixy_
|
|
 |
98.6 |
18 |
PDB header:ribosome
Chain: Y: PDB Molecule:
PDBTitle: structure of rele nuclease bound to the 70s ribosome (postcleavage2 state; part 3 of 4)
|
|
|
|
| 11 | c4mctD_
|
|
 |
98.5 |
23 |
PDB header:toxin
Chain: D: PDB Molecule:killer protein;
PDBTitle: p. vulgaris higba structure, crystal form 1
|
|
|
|
| 12 | c5cw7H_
|
|
 |
98.0 |
13 |
PDB header:toxin
Chain: H: PDB Molecule:plasmid stabilization protein pare;
PDBTitle: crystal structure of the paaa2-pare2 antitoxin-toxin complex
|
|
|
|
| 13 | c5cegB_
|
|
 |
97.9 |
18 |
PDB header:toxin
Chain: B: PDB Molecule:plasmid stabilization system;
PDBTitle: x-ray structure of toxin/anti-toxin complex from mesorhizobium2 opportunistum
|
|
|
|
| 14 | c3kxeB_
|
|
 |
97.7 |
21 |
PDB header:protein binding
Chain: B: PDB Molecule:toxin protein pare-1;
PDBTitle: a conserved mode of protein recognition and binding in a2 pard-pare toxin-antitoxin complex
|
|
|
|
| 15 | c5ja9D_
|
|
 |
95.8 |
18 |
PDB header:toxin
Chain: D: PDB Molecule:toxin higb-2;
PDBTitle: crystal structure of the higb2 toxin in complex with nb6
|
|
|
|
| 16 | c6f8sD_
|
|
 |
95.7 |
17 |
PDB header:toxin
Chain: D: PDB Molecule:putative killer protein;
PDBTitle: toxin-antitoxin complex grata
|
|
|
|
| 17 | d3e9va1
|
|
 |
39.3 |
17 |
Fold:BTG domain-like
Superfamily:BTG domain-like
Family:BTG domain-like |
|
|
|
| 18 | c2lo0B_
|
|
 |
32.6 |
27 |
PDB header:protein binding
Chain: B: PDB Molecule:uncharacterized protein;
PDBTitle: solution structure of the get5 carboxyl domain from a. fumigatus
|
|
|
|
| 19 | d2z15a1
|
|
 |
26.9 |
17 |
Fold:BTG domain-like
Superfamily:BTG domain-like
Family:BTG domain-like |
|
|
|
| 20 | d1khba2
|
|
 |
23.6 |
26 |
Fold:PEP carboxykinase N-terminal domain
Superfamily:PEP carboxykinase N-terminal domain
Family:PEP carboxykinase N-terminal domain |
|
|
|
| 21 | c5j10A_ |
|
not modelled |
20.0 |
32 |
PDB header:de novo protein
Chain: A: PDB Molecule:peptide design 2l4hc2_24;
PDBTitle: de novo design of protein homo-oligomers with modular hydrogen bond2 network-mediated specificity
|
|
|
| 22 | d1s0ua2 |
|
not modelled |
16.0 |
45 |
Fold:Elongation factor/aminomethyltransferase common domain
Superfamily:EF-Tu/eEF-1alpha/eIF2-gamma C-terminal domain
Family:EF-Tu/eEF-1alpha/eIF2-gamma C-terminal domain |
|
|
| 23 | d3bb9a1 |
|
not modelled |
15.7 |
10 |
Fold:Cystatin-like
Superfamily:NTF2-like
Family:SO0125-like |
|
|
| 24 | c3sjaG_ |
|
not modelled |
15.5 |
19 |
PDB header:hydrolase/transport protein
Chain: G: PDB Molecule:golgi to er traffic protein 1;
PDBTitle: crystal structure of s. cerevisiae get3 in the open state in complex2 with get1 cytosolic domain
|
|
|
| 25 | d2qn6a2 |
|
not modelled |
15.3 |
36 |
Fold:Elongation factor/aminomethyltransferase common domain
Superfamily:EF-Tu/eEF-1alpha/eIF2-gamma C-terminal domain
Family:EF-Tu/eEF-1alpha/eIF2-gamma C-terminal domain |
|
|
| 26 | d1kk1a2 |
|
not modelled |
14.4 |
45 |
Fold:Elongation factor/aminomethyltransferase common domain
Superfamily:EF-Tu/eEF-1alpha/eIF2-gamma C-terminal domain
Family:EF-Tu/eEF-1alpha/eIF2-gamma C-terminal domain |
|
|
| 27 | c3sjbC_ |
|
not modelled |
13.4 |
18 |
PDB header:hydrolase/transport protein
Chain: C: PDB Molecule:golgi to er traffic protein 1;
PDBTitle: crystal structure of s. cerevisiae get3 in the open state in complex2 with get1 cytosolic domain
|
|
|
| 28 | c1rb8J_ |
|
not modelled |
13.1 |
40 |
PDB header:virus/dna
Chain: J: PDB Molecule:small core protein;
PDBTitle: the phix174 dna binding protein j in two different capsid2 environments.
|
|
|
| 29 | c2kruA_ |
|
not modelled |
12.0 |
6 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:light-independent protochlorophyllide reductase subunit b;
PDBTitle: solution nmr structure of the pcp_red domain of light-independent2 protochlorophyllide reductase subunit b from chlorobium tepidum.3 northeast structural genomics consortium target ctr69a
|
|
|
| 30 | d2gykb1 |
|
not modelled |
11.7 |
10 |
Fold:His-Me finger endonucleases
Superfamily:His-Me finger endonucleases
Family:HNH-motif |
|
|
| 31 | c5td6A_ |
|
not modelled |
11.4 |
17 |
PDB header:rna binding protein
Chain: A: PDB Molecule:fog-3 protein;
PDBTitle: c. elegans fog-3 btg/tob domain - h47n, c117a
|
|
|
| 32 | c3e56A_ |
|
not modelled |
10.3 |
29 |
PDB header:unknown function
Chain: A: PDB Molecule:putative uncharacterized protein;
PDBTitle: the 2.0 angstrom resolution crystal structure of npr1517, a putative2 heterocyst differentiation inhibitor from nostoc punctiforme
|
|
|
| 33 | c2fh0A_ |
|
not modelled |
10.3 |
11 |
PDB header:unknown function
Chain: A: PDB Molecule:hypothetical 16.0 kda protein in abf2-chl12
PDBTitle: nmr ensemble of the yeast saccharomyces cerevisiae protein2 ymr074cp core region
|
|
|
| 34 | d1eija_ |
|
not modelled |
10.0 |
11 |
Fold:RuvA C-terminal domain-like
Superfamily:Double-stranded DNA-binding domain
Family:Double-stranded DNA-binding domain |
|
|
| 35 | c2qeyA_ |
|
not modelled |
9.9 |
24 |
PDB header:lyase
Chain: A: PDB Molecule:phosphoenolpyruvate carboxykinase, cytosolic [gtp];
PDBTitle: rat cytosolic pepck in complex with gtp
|
|
|
| 36 | c1wmvA_ |
|
not modelled |
9.6 |
21 |
PDB header:oxidoreductase, apoptosis
Chain: A: PDB Molecule:ww domain containing oxidoreductase;
PDBTitle: solution structure of the second ww domain of wwox
|
|
|
| 37 | d2d5ua1 |
|
not modelled |
9.5 |
13 |
Fold:PUG domain-like
Superfamily:PUG domain-like
Family:PUG domain |
|
|
| 38 | c2dogA_ |
|
not modelled |
9.0 |
20 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:probable 16s rrna-processing protein rimm;
PDBTitle: solution structure of the n-terminal domain of rimm from thermus2 thermophilus hb8
|
|
|
| 39 | d2crua1 |
|
not modelled |
7.7 |
11 |
Fold:RuvA C-terminal domain-like
Superfamily:Double-stranded DNA-binding domain
Family:Double-stranded DNA-binding domain |
|
|
| 40 | d2ux0a1 |
|
not modelled |
7.4 |
37 |
Fold:Cystatin-like
Superfamily:NTF2-like
Family:Association domain of calcium/calmodulin-dependent protein kinase type II alpha subunit, CAMK2A |
|
|
| 41 | c6iqcA_ |
|
not modelled |
7.2 |
11 |
PDB header:dna binding protein
Chain: A: PDB Molecule:dna-binding protein sso0352;
PDBTitle: wild-type programmed cell death 5 protein from sulfolobus solfataricus
|
|
|
| 42 | c3d7iB_ |
|
not modelled |
6.9 |
17 |
PDB header:lyase
Chain: B: PDB Molecule:carboxymuconolactone decarboxylase family protein;
PDBTitle: crystal structure of carboxymuconolactone decarboxylase family protein2 possibly involved in oxygen detoxification (1591455) from3 methanococcus jannaschii at 1.75 a resolution
|
|
|
| 43 | c6ms4B_ |
|
not modelled |
6.5 |
12 |
PDB header:translation
Chain: B: PDB Molecule:density-regulated protein;
PDBTitle: crystal structure of the denr-mct-1 complex
|
|
|
| 44 | d1ejxb_ |
|
not modelled |
6.4 |
29 |
Fold:beta-clip
Superfamily:Urease, beta-subunit
Family:Urease, beta-subunit |
|
|
| 45 | c4gicB_ |
|
not modelled |
6.2 |
28 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:histidinol dehydrogenase;
PDBTitle: crystal structure of a putative histidinol dehydrogenase (target psi-2 014034) from methylococcus capsulatus
|
|
|
| 46 | c4z42B_ |
|
not modelled |
5.9 |
14 |
PDB header:hydrolase
Chain: B: PDB Molecule:urease subunit beta;
PDBTitle: crystal structure of urease from yersinia enterocolitica
|
|
|
| 47 | d2jb0b1 |
|
not modelled |
5.8 |
11 |
Fold:His-Me finger endonucleases
Superfamily:His-Me finger endonucleases
Family:HNH-motif |
|
|
| 48 | c2wx4C_ |
|
not modelled |
5.7 |
9 |
PDB header:structural protein
Chain: C: PDB Molecule:decapping protein 1;
PDBTitle: asymmetric trimer of the drosophila melanogaster dcp1 c-2 terminal domain
|
|
|
| 49 | c2l4jA_ |
|
not modelled |
5.6 |
29 |
PDB header:transcription
Chain: A: PDB Molecule:yes-associated protein 2 (yap2);
PDBTitle: yap ww2
|
|
|
| 50 | c2l09A_ |
|
not modelled |
5.5 |
7 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:asr4154 protein;
PDBTitle: solution nmr structure of protein asr4154 from nostoc sp. pcc71202 northeast structural genomics consortium target id nsr143
|
|
|
| 51 | d4ubpb_ |
|
not modelled |
5.5 |
14 |
Fold:beta-clip
Superfamily:Urease, beta-subunit
Family:Urease, beta-subunit |
|
|
| 52 | c2jxnA_ |
|
not modelled |
5.4 |
11 |
PDB header:unknown function
Chain: A: PDB Molecule:uncharacterized protein ymr074c;
PDBTitle: solution structure of s. cerevisiae pdcd5-like protein ymr074cp
|
|
|
| 53 | c5nb9A_ |
|
not modelled |
5.3 |
28 |
PDB header:rna
Chain: A: PDB Molecule:rna chaperone proq;
PDBTitle: structure of the n-terminal domain of the escherichia coli proq rna2 binding protein
|
|
|