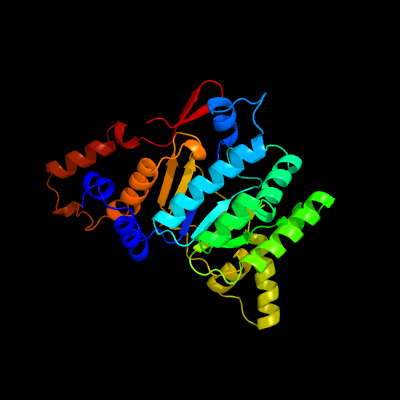
| 1 | c4cmxB_
|
|
 |
100.0 |
100 |
PDB header:nuclear protein
Chain: B: PDB Molecule:rv3378c;
PDBTitle: crystal structure of rv3378c
|
|
|
|
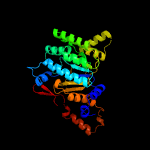
| 2 | c6acsA_
|
|
 |
98.9 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:ditrans,polycis-undecaprenyl-diphosphate synthase ((2e,6e)-
PDBTitle: poly-cis-prenyltransferase
|
|
|
|
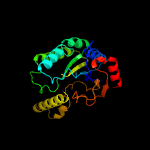
| 3 | c4h8eA_
|
|
 |
98.6 |
13 |
PDB header:transferase/transferase inhibitor
Chain: A: PDB Molecule:undecaprenyl pyrophosphate synthase;
PDBTitle: structure of s. aureus undecaprenyl diphosphate synthase in complex2 with fpp and sulfate
|
|
|
|
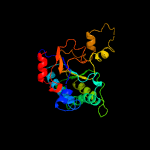
| 4 | c2vg2C_
|
|
 |
98.5 |
15 |
PDB header:transferase
Chain: C: PDB Molecule:undecaprenyl pyrophosphate synthetase;
PDBTitle: rv2361 with ipp
|
|
|
|
| 5 | c5hc7A_
|
|
 |
98.5 |
14 |
PDB header:transferase/transferase inhibitor
Chain: A: PDB Molecule:prenyltransference for protein;
PDBTitle: crystal structure of lavandulyl diphosphate synthase from lavandula x2 intermedia in complex with s-thiolo-isopentenyldiphosphate
|
|
|
|
| 6 | c5hxpA_
|
|
 |
98.4 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:(2z,6z)-farnesyl diphosphate synthase, chloroplastic;
PDBTitle: crystal structure of z,z-farnesyl diphosphate synthase (d71m, e75a and2 h103y mutants) complexed with ipp
|
|
|
|
| 7 | c4q9mA_
|
|
 |
98.4 |
16 |
PDB header:transferase/transferase inhibitor
Chain: A: PDB Molecule:isoprenyl transferase;
PDBTitle: crystal structure of upps in complex with fpp and an allosteric2 inhibitor
|
|
|
|
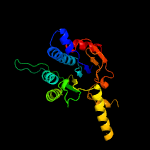
| 8 | d1f75a_
|
|
 |
98.3 |
16 |
Fold:Undecaprenyl diphosphate synthase
Superfamily:Undecaprenyl diphosphate synthase
Family:Undecaprenyl diphosphate synthase |
|
|
|
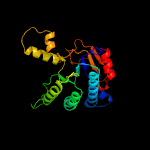
| 9 | d1ueha_
|
|
 |
98.3 |
17 |
Fold:Undecaprenyl diphosphate synthase
Superfamily:Undecaprenyl diphosphate synthase
Family:Undecaprenyl diphosphate synthase |
|
|
|
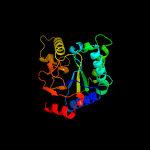
| 10 | c3ugsB_
|
|
 |
98.3 |
15 |
PDB header:transferase
Chain: B: PDB Molecule:undecaprenyl pyrophosphate synthase;
PDBTitle: crystal structure of a probable undecaprenyl diphosphate synthase2 (upps) from campylobacter jejuni
|
|
|
|
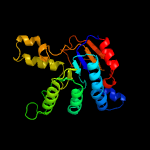
| 11 | c1jp3A_
|
|
 |
98.2 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:undecaprenyl pyrophosphate synthase;
PDBTitle: structure of e.coli undecaprenyl pyrophosphate synthase
|
|
|
|
| 12 | c2d2rA_
|
|
 |
98.2 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:undecaprenyl pyrophosphate synthase;
PDBTitle: crystal structure of helicobacter pylori undecaprenyl pyrophosphate2 synthase
|
|
|
|
| 13 | c2vfwB_
|
|
 |
98.1 |
19 |
PDB header:transferase
Chain: B: PDB Molecule:short-chain z-isoprenyl diphosphate synthetase;
PDBTitle: rv1086 native
|
|
|
|
| 14 | c5xk9F_
|
|
 |
97.7 |
18 |
PDB header:transferase
Chain: F: PDB Molecule:undecaprenyl diphosphate synthase;
PDBTitle: crystal structure of isosesquilavandulyl diphosphate synthase from2 streptomyces sp. strain cnh-189 in complex with gspp and dmapp
|
|
|
|
| 15 | c5gukA_
|
|
 |
97.1 |
19 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:cyclolavandulyl diphosphate synthase;
PDBTitle: crystal structure of apo form of cyclolavandulyl diphosphate synthase2 (clds) from streptomyces sp. cl190
|
|
|
|
| 16 | d1jhfa1
|
|
 |
68.7 |
31 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:LexA repressor, N-terminal DNA-binding domain |
|
|
|
| 17 | c6jcnB_
|
|
 |
58.3 |
11 |
PDB header:transferase
Chain: B: PDB Molecule:dehydrodolichyl diphosphate synthase complex subunit nus1;
PDBTitle: yeast dehydrodolichyl diphosphate synthase complex subunit nus1
|
|
|
|
| 18 | c4kvjA_
|
|
 |
45.1 |
30 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:fatty acid alpha-oxidase;
PDBTitle: crystal structure of oryza sativa fatty acid alpha-dioxygenase with2 hydrogen peroxide
|
|
|
|
| 19 | c5iq5A_
|
|
 |
44.4 |
20 |
PDB header:viral protein
Chain: A: PDB Molecule:macro domain;
PDBTitle: nmr solution structure of mayaro virus macro domain
|
|
|
|
| 20 | c2kmuA_
|
|
 |
38.9 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:atp-dependent dna helicase q4;
PDBTitle: recql4 amino-terminal domain
|
|
|
|
| 21 | d1k8ba_ |
|
not modelled |
37.9 |
19 |
Fold:Ribosome binding domain-like
Superfamily:Translation initiation factor 2 beta, aIF2beta, N-terminal domain
Family:Translation initiation factor 2 beta, aIF2beta, N-terminal domain |
|
|
| 22 | d1xo1a2 |
|
not modelled |
31.9 |
17 |
Fold:PIN domain-like
Superfamily:PIN domain-like
Family:5' to 3' exonuclease catalytic domain |
|
|
| 23 | d1qwga_ |
|
not modelled |
30.7 |
24 |
Fold:TIM beta/alpha-barrel
Superfamily:(2r)-phospho-3-sulfolactate synthase ComA
Family:(2r)-phospho-3-sulfolactate synthase ComA |
|
|
| 24 | c6b1zA_ |
|
not modelled |
30.7 |
21 |
PDB header:ligase
Chain: A: PDB Molecule:glutamate--trna ligase;
PDBTitle: crystal structure of glutamate-trna synthetase from elizabethkingia2 anophelis
|
|
|
| 25 | c2e9hA_ |
|
not modelled |
30.2 |
30 |
PDB header:translation
Chain: A: PDB Molecule:eukaryotic translation initiation factor 5;
PDBTitle: solution structure of the eif-5_eif-2b domain from human2 eukaryotic translation initiation factor 5
|
|
|
| 26 | c5hr4J_ |
|
not modelled |
26.9 |
24 |
PDB header:hydrolase/dna
Chain: J: PDB Molecule:mmei;
PDBTitle: structure of type iil restriction-modification enzyme mmei in complex2 with dna has implications for engineering of new specificities
|
|
|
| 27 | c3ff4A_ |
|
not modelled |
26.5 |
14 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of uncharacterized protein chu_1412
|
|
|
| 28 | d1cvua1 |
|
not modelled |
25.7 |
10 |
Fold:Heme-dependent peroxidases
Superfamily:Heme-dependent peroxidases
Family:Myeloperoxidase-like |
|
|
| 29 | c1ddxA_ |
|
not modelled |
25.6 |
10 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:protein (prostaglandin h2 synthase-2);
PDBTitle: crystal structure of a mixture of arachidonic acid and prostaglandin2 bound to the cyclooxygenase active site of cox-2: prostaglandin3 structure
|
|
|
| 30 | c5bthA_ |
|
not modelled |
24.4 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:decapping nuclease rai1;
PDBTitle: crystal structure of candida albicans rai1
|
|
|
| 31 | c1oheA_ |
|
not modelled |
24.0 |
20 |
PDB header:hydrolase
Chain: A: PDB Molecule:cdc14b2 phosphatase;
PDBTitle: structure of cdc14b phosphatase with a peptide ligand
|
|
|
| 32 | c1pggB_ |
|
not modelled |
23.6 |
17 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:prostaglandin h2 synthase-1;
PDBTitle: prostaglandin h2 synthase-1 complexed with 1-(4-iodobenzoyl)-5-2 methoxy-2-methylindole-3-acetic acid (iodoindomethacin), trans model
|
|
|
| 33 | c1ht8B_ |
|
not modelled |
23.5 |
17 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:prostaglandin h2 synthase-1;
PDBTitle: the 2.7 angstrom resolution model of ovine cox-1 complexed with2 alclofenac
|
|
|
| 34 | d1bdga2 |
|
not modelled |
22.9 |
14 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:Hexokinase |
|
|
| 35 | c3pghD_ |
|
not modelled |
22.6 |
10 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:cyclooxygenase-2;
PDBTitle: cyclooxygenase-2 (prostaglandin synthase-2) complexed with a non-2 selective inhibitor, flurbiprofen
|
|
|
| 36 | c6md3F_ |
|
not modelled |
22.4 |
17 |
PDB header:hydrolase/rna
Chain: F: PDB Molecule:rrp44p homologue;
PDBTitle: structure of t. brucei rrp44 pin domain
|
|
|
| 37 | d1q4ga1 |
|
not modelled |
22.1 |
17 |
Fold:Heme-dependent peroxidases
Superfamily:Heme-dependent peroxidases
Family:Myeloperoxidase-like |
|
|
| 38 | c1ig8A_ |
|
not modelled |
21.7 |
24 |
PDB header:transferase
Chain: A: PDB Molecule:hexokinase pii;
PDBTitle: crystal structure of yeast hexokinase pii with the correct2 amino acid sequence
|
|
|
| 39 | c4zwjA_ |
|
not modelled |
21.0 |
26 |
PDB header:signaling protein
Chain: A: PDB Molecule:chimera protein of human rhodopsin, mouse s-arrestin, and
PDBTitle: crystal structure of rhodopsin bound to arrestin by femtosecond x-ray2 laser
|
|
|
| 40 | c3gebC_ |
|
not modelled |
20.8 |
25 |
PDB header:hydrolase
Chain: C: PDB Molecule:eyes absent homolog 2;
PDBTitle: crystal structure of edeya2
|
|
|
| 41 | c2oyuP_ |
|
not modelled |
20.5 |
17 |
PDB header:oxidoreductase
Chain: P: PDB Molecule:prostaglandin g/h synthase 1;
PDBTitle: indomethacin-(s)-alpha-ethyl-ethanolamide bound to cyclooxygenase-1
|
|
|
| 42 | d2bgxa1 |
|
not modelled |
20.4 |
37 |
Fold:PGBD-like
Superfamily:PGBD-like
Family:Peptidoglycan binding domain, PGBD |
|
|
| 43 | c5uqyB_ |
|
not modelled |
20.1 |
20 |
PDB header:viral protein/immune system
Chain: B: PDB Molecule:envelope glycoprotein gp2;
PDBTitle: crystal structure of marburg virus gp in complex with the human2 survivor antibody mr78
|
|
|
| 44 | c6d6rK_ |
|
not modelled |
19.9 |
20 |
PDB header:hydrolase
Chain: K: PDB Molecule:exosome complex exonuclease rrp44;
PDBTitle: human nuclear exosome-mtr4 rna complex - composite map after focused2 reconstruction
|
|
|
| 45 | d2npta1 |
|
not modelled |
19.3 |
15 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:CAD & PB1 domains
Family:PB1 domain |
|
|
| 46 | c6ajjA_ |
|
not modelled |
19.1 |
26 |
PDB header:membrane protein, hydrolase
Chain: A: PDB Molecule:drug exporters of the rnd superfamily-like protein,
PDBTitle: crystal structure of mycolic acid transporter mmpl3 from mycobacterium2 smegmatis complexed with ica38
|
|
|
| 47 | c1jpeA_ |
|
not modelled |
18.6 |
20 |
PDB header:electron transport
Chain: A: PDB Molecule:dsbd-alpha;
PDBTitle: crystal structure of dsbd-alpha; the n-terminal domain of2 dsbd
|
|
|
| 48 | d1ig8a2 |
|
not modelled |
18.5 |
24 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:Hexokinase |
|
|
| 49 | d1iuka_ |
|
not modelled |
18.4 |
17 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:CoA-binding domain |
|
|
| 50 | c2of5A_ |
|
not modelled |
18.3 |
21 |
PDB header:apoptosis
Chain: A: PDB Molecule:death domain-containing protein cradd;
PDBTitle: oligomeric death domain complex
|
|
|
| 51 | c4pv6E_ |
|
not modelled |
18.1 |
19 |
PDB header:transferase
Chain: E: PDB Molecule:n-terminal acetyltransferase complex subunit [ard1];
PDBTitle: crystal structure analysis of ard1 from thermoplasma volcanium
|
|
|
| 52 | c4zwjC_ |
|
not modelled |
17.5 |
26 |
PDB header:signaling protein
Chain: C: PDB Molecule:chimera protein of human rhodopsin, mouse s-arrestin, and
PDBTitle: crystal structure of rhodopsin bound to arrestin by femtosecond x-ray2 laser
|
|
|
| 53 | d2qtva4 |
|
not modelled |
17.3 |
27 |
Fold:Gelsolin-like
Superfamily:C-terminal, gelsolin-like domain of Sec23/24
Family:C-terminal, gelsolin-like domain of Sec23/24 |
|
|
| 54 | d1iuqa_ |
|
not modelled |
17.2 |
12 |
Fold:Glycerol-3-phosphate (1)-acyltransferase
Superfamily:Glycerol-3-phosphate (1)-acyltransferase
Family:Glycerol-3-phosphate (1)-acyltransferase |
|
|
| 55 | d1u83a_ |
|
not modelled |
17.2 |
24 |
Fold:TIM beta/alpha-barrel
Superfamily:(2r)-phospho-3-sulfolactate synthase ComA
Family:(2r)-phospho-3-sulfolactate synthase ComA |
|
|
| 56 | c1u83A_ |
|
not modelled |
17.2 |
24 |
PDB header:lyase
Chain: A: PDB Molecule:phosphosulfolactate synthase;
PDBTitle: psl synthase from bacillus subtilis
|
|
|
| 57 | c4p9eA_ |
|
not modelled |
16.8 |
27 |
PDB header:hydrolase
Chain: A: PDB Molecule:deoxycytidylate deaminase;
PDBTitle: crystal structure of dcmp deaminase from the cyanophage s-tim5 in apo2 form
|
|
|
| 58 | c3al0C_ |
|
not modelled |
16.7 |
16 |
PDB header:ligase/rna
Chain: C: PDB Molecule:glutamyl-trna(gln) amidotransferase subunit c,linker,
PDBTitle: crystal structure of the glutamine transamidosome from thermotoga2 maritima in the glutamylation state.
|
|
|
| 59 | d1l6pa_ |
|
not modelled |
16.6 |
20 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Thiol:disulfide interchange protein DsbD, N-terminal domain (DsbD-alpha)
Family:Thiol:disulfide interchange protein DsbD, N-terminal domain (DsbD-alpha) |
|
|
| 60 | c3zddA_ |
|
not modelled |
16.2 |
17 |
PDB header:hydrolase/dna
Chain: A: PDB Molecule:protein xni;
PDBTitle: structure of e. coli exoix in complex with the palindromic 5ov62 oligonucleotide and potassium
|
|
|
| 61 | c3j6vJ_ |
|
not modelled |
15.3 |
30 |
PDB header:ribosome
Chain: J: PDB Molecule:28s ribosomal protein s10, mitochondrial;
PDBTitle: cryo-em structure of the small subunit of the mammalian mitochondrial2 ribosome
|
|
|
| 62 | c2x7bA_ |
|
not modelled |
15.2 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:n-acetyltransferase sso0209;
PDBTitle: crystal structure of the n-terminal acetylase ard1 from2 sulfolobus solfataricus p2
|
|
|
| 63 | d1l0oc_ |
|
not modelled |
14.9 |
20 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Sigma3 and sigma4 domains of RNA polymerase sigma factors
Family:Sigma4 domain |
|
|
| 64 | c1l0oC_ |
|
not modelled |
14.9 |
20 |
PDB header:protein binding
Chain: C: PDB Molecule:sigma factor;
PDBTitle: crystal structure of the bacillus stearothermophilus anti-2 sigma factor spoiiab with the sporulation sigma factor3 sigmaf
|
|
|
| 65 | c2fb6A_ |
|
not modelled |
14.7 |
13 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:conserved hypothetical protein;
PDBTitle: structure of conserved protein of unknown function bt1422 from2 bacteroides thetaiotaomicron
|
|
|
| 66 | c4bqeA_ |
|
not modelled |
14.5 |
23 |
PDB header:transferase
Chain: A: PDB Molecule:alpha-glucan phosphorylase 2,4-glucan phosphorylase;
PDBTitle: arabidopsis thaliana cytosolic alpha-1,4-glucan phosphorylase (phs2)
|
|
|
| 67 | c2mxtA_ |
|
not modelled |
14.5 |
13 |
PDB header:splicing
Chain: A: PDB Molecule:heterogeneous nuclear ribonucleoprotein q;
PDBTitle: nmr structure of the acidic domain of syncrip (hnrnpq)
|
|
|
| 68 | d2bcqa2 |
|
not modelled |
13.9 |
21 |
Fold:SAM domain-like
Superfamily:PsbU/PolX domain-like
Family:DNA polymerase beta-like, second domain |
|
|
| 69 | d1o98a1 |
|
not modelled |
13.8 |
18 |
Fold:2,3-Bisphosphoglycerate-independent phosphoglycerate mutase, substrate-binding domain
Superfamily:2,3-Bisphosphoglycerate-independent phosphoglycerate mutase, substrate-binding domain
Family:2,3-Bisphosphoglycerate-independent phosphoglycerate mutase, substrate-binding domain |
|
|
| 70 | c5y26A_ |
|
not modelled |
13.6 |
27 |
PDB header:dna binding protein
Chain: A: PDB Molecule:dna polymerase epsilon subunit d;
PDBTitle: crystal structure of native dpb4-dpb3
|
|
|
| 71 | c4f0qC_ |
|
not modelled |
13.5 |
18 |
PDB header:hydrolase
Chain: C: PDB Molecule:restriction endonuclease;
PDBTitle: mspji restriction endonuclease - p21 form
|
|
|
| 72 | c4f0qA_ |
|
not modelled |
13.5 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:restriction endonuclease;
PDBTitle: mspji restriction endonuclease - p21 form
|
|
|
| 73 | c3d7iB_ |
|
not modelled |
13.4 |
17 |
PDB header:lyase
Chain: B: PDB Molecule:carboxymuconolactone decarboxylase family protein;
PDBTitle: crystal structure of carboxymuconolactone decarboxylase family protein2 possibly involved in oxygen detoxification (1591455) from3 methanococcus jannaschii at 1.75 a resolution
|
|
|
| 74 | c2a5wC_ |
|
not modelled |
13.2 |
9 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:sulfite reductase, desulfoviridin-type subunit gamma
PDBTitle: crystal structure of the oxidized gamma-subunit of the dissimilatory2 sulfite reductase (dsrc) from archaeoglobus fulgidus
|
|
|
| 75 | c2k0rA_ |
|
not modelled |
13.1 |
33 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:thiol:disulfide interchange protein dsbd;
PDBTitle: solution structure of the c103s mutant of the n-terminal2 domain of dsbd from neisseria meningitidis
|
|
|
| 76 | c5kzoA_ |
|
not modelled |
13.0 |
44 |
PDB header:transcription
Chain: A: PDB Molecule:neurogenic locus notch homolog protein 1;
PDBTitle: notch1 transmembrane and associated juxtamembrane segment
|
|
|
| 77 | c6c95B_ |
|
not modelled |
12.7 |
12 |
PDB header:transferase
Chain: B: PDB Molecule:n-alpha-acetyltransferase 10;
PDBTitle: the human nata (naa10/naa15) amino-terminal acetyltransferase complex2 bound to hypk
|
|
|
| 78 | d1stza1 |
|
not modelled |
12.7 |
15 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:Heat-inducible transcription repressor HrcA, N-terminal domain |
|
|
| 79 | c3jsoA_ |
|
not modelled |
12.4 |
31 |
PDB header:hydrolase/dna
Chain: A: PDB Molecule:lexa repressor;
PDBTitle: classic protein with a new twist: crystal structure of a lexa2 repressor dna complex
|
|
|
| 80 | d1ygha_ |
|
not modelled |
12.2 |
9 |
Fold:Acyl-CoA N-acyltransferases (Nat)
Superfamily:Acyl-CoA N-acyltransferases (Nat)
Family:N-acetyl transferase, NAT |
|
|
| 81 | d2icwg1 |
|
not modelled |
11.9 |
23 |
Fold:Superantigen MAM
Superfamily:Superantigen MAM
Family:Superantigen MAM |
|
|
| 82 | c2jpmA_ |
|
not modelled |
11.5 |
15 |
PDB header:antimicrobial protein
Chain: A: PDB Molecule:bacteriocin lactococcin-g subunit beta;
PDBTitle: lactococcin g-b in tfe
|
|
|
| 83 | c3qbuD_ |
|
not modelled |
11.4 |
13 |
PDB header:hydrolase
Chain: D: PDB Molecule:putative uncharacterized protein;
PDBTitle: crystal structure of putative peptidoglycan deactelyase (hp0310) from2 helicobacter pylori
|
|
|
| 84 | c2n6vA_ |
|
not modelled |
11.4 |
25 |
PDB header:unknown function
Chain: A: PDB Molecule:astexin3;
PDBTitle: solution study of astexin3
|
|
|
| 85 | c1yx3A_ |
|
not modelled |
11.3 |
12 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein dsrc;
PDBTitle: nmr structure of allochromatium vinosum dsrc: northeast2 structural genomics consortium target op4
|
|
|
| 86 | c4giwB_ |
|
not modelled |
11.3 |
24 |
PDB header:signaling protein
Chain: B: PDB Molecule:run and sh3 domain-containing protein 1;
PDBTitle: crystal structure of the run domain of human nesca
|
|
|
| 87 | c4im4F_ |
|
not modelled |
11.1 |
13 |
PDB header:hydrolase
Chain: F: PDB Molecule:endoglucanase e;
PDBTitle: multifunctional cellulase, xylanase, mannanase
|
|
|
| 88 | c3wo6A_ |
|
not modelled |
10.9 |
13 |
PDB header:membrane protein
Chain: A: PDB Molecule:membrane protein insertase yidc 2;
PDBTitle: crystal structure of yidc from bacillus halodurans (form i)
|
|
|
| 89 | d2iw0a1 |
|
not modelled |
10.8 |
15 |
Fold:7-stranded beta/alpha barrel
Superfamily:Glycoside hydrolase/deacetylase
Family:NodB-like polysaccharide deacetylase |
|
|
| 90 | c4e5gB_ |
|
not modelled |
10.8 |
20 |
PDB header:viral protein, transcription
Chain: B: PDB Molecule:polymerase protein pa;
PDBTitle: crystal structure of avian influenza virus pan bound to compound 2
|
|
|
| 91 | c4oc8A_ |
|
not modelled |
10.7 |
23 |
PDB header:hydrolase
Chain: A: PDB Molecule:restriction endonuclease aspbhi;
PDBTitle: dna modification-dependent restriction endonuclease aspbhi
|
|
|
| 92 | c2jpkA_ |
|
not modelled |
10.7 |
15 |
PDB header:antimicrobial protein
Chain: A: PDB Molecule:bacteriocin lactococcin-g subunit beta;
PDBTitle: lactococcin g-b in dpc
|
|
|
| 93 | c4griB_ |
|
not modelled |
10.6 |
18 |
PDB header:ligase
Chain: B: PDB Molecule:glutamate--trna ligase;
PDBTitle: crystal structure of a glutamyl-trna synthetase glurs from borrelia2 burgdorferi bound to glutamic acid and zinc
|
|
|
| 94 | c4hhsA_ |
|
not modelled |
10.6 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:alpha-dioxygenase;
PDBTitle: crystal structure of fatty acid alpha-dioxygenase (arabidopsis2 thaliana)
|
|
|
| 95 | c5ccuA_ |
|
not modelled |
10.6 |
24 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative secreted endoglycosylceramidase;
PDBTitle: crystal structure of endoglycoceramidase i from rhodococ-cus equi
|
|
|
| 96 | c3gqeA_ |
|
not modelled |
10.5 |
13 |
PDB header:viral protein
Chain: A: PDB Molecule:non-structural protein 3;
PDBTitle: crystal structure of macro domain of venezuelan equine encephalitis2 virus
|
|
|
| 97 | c6mzdC_ |
|
not modelled |
10.4 |
18 |
PDB header:transcription
Chain: C: PDB Molecule:transcription initiation factor tfiid subunit 3;
PDBTitle: human tfiid lobe a canonical
|
|
|
| 98 | c2m8fA_ |
|
not modelled |
10.3 |
25 |
PDB header:unknown function
Chain: A: PDB Molecule:astexin3;
PDBTitle: structure of lasso peptide astexin3
|
|
|
| 99 | c1kbiB_ |
|
not modelled |
10.3 |
28 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:cytochrome b2;
PDBTitle: crystallographic study of the recombinant flavin-binding domain of2 baker's yeast flavocytochrome b2: comparison with the intact wild-3 type enzyme
|
|
|