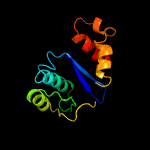
1 d1musa_
98.0
17
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: Transposase inhibitor (Tn5 transposase)
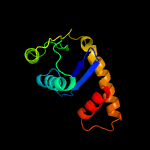
2 d1b7ea_
97.2
20
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: Transposase inhibitor (Tn5 transposase)
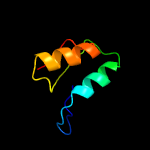
3 d1asua_
91.0
17
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: Retroviral integrase, catalytic domain
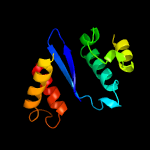
4 d1cxqa_
78.3
19
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: Retroviral integrase, catalytic domain
5 c5cz1B_
64.5
12
PDB header: hydrolaseChain: B: PDB Molecule: integrase;PDBTitle: crystal structure of the catalytic core domain of mmtv integrase
6 c3nf9A_
62.3
18
PDB header: hydrolase/hydrolase inhibitorChain: A: PDB Molecule: integrase;PDBTitle: structural basis for a new mechanism of inhibition of hiv integrase2 identified by fragment screening and structure based design
7 c1c0mA_
50.3
20
PDB header: transferaseChain: A: PDB Molecule: protein (integrase);PDBTitle: crystal structure of rsv two-domain integrase
8 c4mq3A_
49.2
14
PDB header: viral proteinChain: A: PDB Molecule: integrase;PDBTitle: the 1.1 angstrom structure of catalytic core domain of fiv integrase
9 c5j9wB_
43.5
14
PDB header: transferaseChain: B: PDB Molecule: chromatin modification-related protein eaf6;PDBTitle: crystal structure of the nua4 core complex
10 d1h5ya_
31.3
19
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: Histidine biosynthesis enzymes
11 c3kksB_
29.4
13
PDB header: dna binding proteinChain: B: PDB Molecule: integrase;PDBTitle: crystal structure of catalytic core domain of biv integrase in crystal2 form ii
12 c3j6vI_
28.1
20
PDB header: ribosomeChain: I: PDB Molecule: 28s ribosomal protein s9, mitochondrial;PDBTitle: cryo-em structure of the small subunit of the mammalian mitochondrial2 ribosome
13 d1exqa_
25.6
18
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: Retroviral integrase, catalytic domain
14 c3jcaE_
23.5
12
PDB header: viral proteinChain: E: PDB Molecule: integrase;PDBTitle: core model of the mouse mammary tumor virus intasome
15 c3tsmB_
21.3
16
PDB header: lyaseChain: B: PDB Molecule: indole-3-glycerol phosphate synthase;PDBTitle: crystal structure of indole-3-glycerol phosphate synthase from2 brucella melitensis
16 c5u1cD_
19.5
24
PDB header: viral proteinChain: D: PDB Molecule: hiv-1 integrase, sso7d chimera;PDBTitle: structure of tetrameric hiv-1 strand transfer complex intasome
17 c3qjaA_
18.4
18
PDB header: lyaseChain: A: PDB Molecule: indole-3-glycerol phosphate synthase;PDBTitle: crystal structure of the mycobacterium tuberculosis indole-3-glycerol2 phosphate synthase (trpc) in apo form
18 c3hpgC_
15.4
12
PDB header: transferaseChain: C: PDB Molecule: integrase;PDBTitle: visna virus integrase (residues 1-219) in complex with ledgf2 ibd: examples of open integrase dimer-dimer interfaces
19 c5ejkG_
15.4
18
PDB header: transferase/dnaChain: G: PDB Molecule: gag-pro-pol polyprotein;PDBTitle: crystal structure of the rous sarcoma virus intasome
20 c3f9kV_
15.3
13
PDB header: viral protein, recombinationChain: V: PDB Molecule: integrase;PDBTitle: two domain fragment of hiv-2 integrase in complex with ledgf ibd
21 d1ka9f_
not modelled
15.1
12
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: Histidine biosynthesis enzymes
22 c5u1cA_
not modelled
14.5
17
PDB header: viral proteinChain: A: PDB Molecule: hiv-1 integrase, sso7d chimera;PDBTitle: structure of tetrameric hiv-1 strand transfer complex intasome
23 d2h1ta1
not modelled
14.3
24
Fold: Spiral beta-rollSuperfamily: PA1994-likeFamily: PA1994-like
24 c6orjA_
not modelled
14.3
20
PDB header: viral proteinChain: A: PDB Molecule: phikz164;PDBTitle: central spike of phikz phage tail
25 c2etjA_
not modelled
13.7
16
PDB header: hydrolaseChain: A: PDB Molecule: ribonuclease hii;PDBTitle: crystal structure of ribonuclease hii (ec 3.1.26.4) (rnase hii)2 (tm0915) from thermotoga maritima at 1.74 a resolution
26 d2etja1
not modelled
13.7
16
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: Ribonuclease H
27 c1k6yB_
not modelled
13.6
21
PDB header: transferaseChain: B: PDB Molecule: integrase;PDBTitle: crystal structure of a two-domain fragment of hiv-1 integrase
28 d1c0ma2
not modelled
13.6
16
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: Retroviral integrase, catalytic domain
29 c6bmaA_
not modelled
13.6
18
PDB header: lyaseChain: A: PDB Molecule: indole-3-glycerol phosphate synthase;PDBTitle: the crystal structure of indole-3-glycerol phosphate synthase from2 campylobacter jejuni subsp. jejuni nctc 11168
30 d1c6va_
not modelled
13.2
19
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: Retroviral integrase, catalytic domain
31 d1bcoa2
not modelled
13.1
12
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: mu transposase, core domain
32 d1a53a_
not modelled
11.1
24
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: Tryptophan biosynthesis enzymes
33 c5uncB_
not modelled
8.8
14
PDB header: isomeraseChain: B: PDB Molecule: phosphoenolpyruvate phosphomutase;PDBTitle: the crystal structure of phosphoenolpyruvate phosphomutase from2 streptomyces platensis subsp. rosaceus
34 c4v1af_
not modelled
8.2
17
PDB header: ribosomeChain: F: PDB Molecule: PDBTitle: structure of the large subunit of the mammalian mitoribosome, part 22 of 2
35 c2wcvI_
not modelled
7.5
12
PDB header: isomeraseChain: I: PDB Molecule: l-fucose mutarotase;PDBTitle: crystal structure of bacterial fucu
36 c5mlpA_
not modelled
7.3
21
PDB header: ligaseChain: A: PDB Molecule: uncharacterized protein;PDBTitle: structure of cdps from rickettsiella grylli
37 c5ooma_
not modelled
7.2
14
PDB header: ribosomeChain: A: PDB Molecule: 16s ribosomal rna;PDBTitle: structure of a native assembly intermediate of the human mitochondrial2 ribosome with unfolded interfacial rrna
38 c3mvkA_
not modelled
7.0
20
PDB header: isomeraseChain: A: PDB Molecule: protein fucu;PDBTitle: the crystal structure of fucu from bifidobacterium longum to 1.65a
39 d1zuna1
not modelled
6.8
8
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Adenine nucleotide alpha hydrolases-likeFamily: PAPS reductase-like
40 c1bcoA_
not modelled
6.7
14
PDB header: transposaseChain: A: PDB Molecule: bacteriophage mu transposase;PDBTitle: bacteriophage mu transposase core domain
41 c3e7nB_
not modelled
6.4
28
PDB header: transport proteinChain: B: PDB Molecule: d-ribose high-affinity transport system;PDBTitle: crystal structure of d-ribose high-affinity transport system from2 salmonella typhimurium lt2
42 c5vliC_
not modelled
6.2
25
PDB header: viral protein/de novo proteinChain: C: PDB Molecule: computationally designed peptide hb1.6928.2.3;PDBTitle: computationally designed inhibitor peptide hb1.6928.2.3 in complex2 with influenza hemagglutinin (a/puertorico/8/1934)
43 c3p13B_
not modelled
6.2
20
PDB header: isomeraseChain: B: PDB Molecule: d-ribose pyranase;PDBTitle: complex structure of d-ribose pyranase sa240 with d-ribose
44 c1zunA_
not modelled
6.2
8
PDB header: transferaseChain: A: PDB Molecule: sulfate adenylyltransferase subunit 2;PDBTitle: crystal structure of a gtp-regulated atp sulfurylase2 heterodimer from pseudomonas syringae
45 c4a34L_
not modelled
6.1
20
PDB header: isomeraseChain: L: PDB Molecule: rbsd/fucu transport protein family protein;PDBTitle: crystal structure of the fucose mutarotase in complex with2 l-fucose from streptococcus pneumoniae
46 c2oq2B_
not modelled
5.9
6
PDB header: oxidoreductaseChain: B: PDB Molecule: phosphoadenosine phosphosulfate reductase;PDBTitle: crystal structure of yeast paps reductase with pap, a product complex
47 d1ogda_
not modelled
5.5
28
Fold: RbsD-likeSuperfamily: RbsD-likeFamily: RbsD-like
48 d2ob5a1
not modelled
5.4
20
Fold: RbsD-likeSuperfamily: RbsD-likeFamily: RbsD-like
49 c3hosA_
not modelled
5.4
14
PDB header: transferase, dna binding protein/dnaChain: A: PDB Molecule: transposable element mariner, complete cds;PDBTitle: crystal structure of the mariner mos1 paired end complex with mg
50 c6hlwB_
not modelled
5.1
15
PDB header: viral proteinChain: B: PDB Molecule: genome polyprotein;PDBTitle: crystal structure of human acbd3 gold domain in complex with 3a2 protein of enterovirus-a71 (fusion protein)