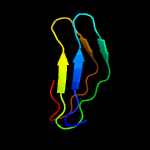
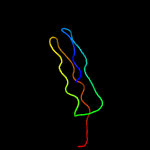
1 c5xfsA_
99.3
47
PDB header: protein transportChain: A: PDB Molecule: pe family protein pe8;PDBTitle: crystal structure of pe8-ppe15 in complex with espg5 from m.2 tuberculosis
2 c2g38A_
99.3
32
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: pe family protein;PDBTitle: a pe/ppe protein complex from mycobacterium tuberculosis
3 d2g38a1
99.3
32
Fold: Ferritin-likeSuperfamily: PE/PPE dimer-likeFamily: PE
4 c1k7qA_
98.0
24
PDB header: hydrolaseChain: A: PDB Molecule: secreted protease c;PDBTitle: prtc from erwinia chrysanthemi: e189a mutant
5 d1kapp1
98.0
19
Fold: Single-stranded right-handed beta-helixSuperfamily: beta-RollFamily: Serralysin-like metalloprotease, C-terminal domain
6 c2ml3A_
97.8
21
PDB header: isomeraseChain: A: PDB Molecule: poly(beta-d-mannuronate) c5 epimerase 6;PDBTitle: solution structure of alge6r3 subunit from the azotobacter vinelandii2 mannuronan c5-epimerase
7 c2agmA_
97.8
27
PDB header: isomeraseChain: A: PDB Molecule: poly(beta-d-mannuronate) c5 epimerase 4;PDBTitle: solution structure of the r-module from alge4
8 c2ml2A_
97.8
21
PDB header: isomeraseChain: A: PDB Molecule: poly(beta-d-mannuronate) c5 epimerase 6;PDBTitle: solution structure of alge6r2 subunit from the azotobacter vinelandii2 mannuronan c5-epimerase
9 d1k7ia1
97.8
28
Fold: Single-stranded right-handed beta-helixSuperfamily: beta-RollFamily: Serralysin-like metalloprotease, C-terminal domain
10 c2zj6A_
97.8
26
PDB header: hydrolaseChain: A: PDB Molecule: lipase;PDBTitle: crystal structure of d337a mutant of pseudomonas sp. mis38 lipase
11 c1jiwP_
97.6
28
PDB header: hydrolase/hyrolase inhibitorChain: P: PDB Molecule: alkaline metalloproteinase;PDBTitle: crystal structure of the apr-aprin complex
12 c1om8A_
97.4
25
PDB header: hydrolaseChain: A: PDB Molecule: serralysin;PDBTitle: crystal structure of a cold adapted alkaline protease from pseudomonas2 tac ii 18, co-crystallyzed with 10 mm edta
13 c2qubG_
97.3
17
PDB header: hydrolaseChain: G: PDB Molecule: extracellular lipase;PDBTitle: crystal structure of extracellular lipase lipa from serratia2 marcescens
14 c1satA_
97.2
25
PDB header: hydrolase (serine protease)Chain: A: PDB Molecule: serratia protease;PDBTitle: crystal structure of the 50 kda metallo protease from s.2 marcescens
15 d1g9ka1
96.7
19
Fold: Single-stranded right-handed beta-helixSuperfamily: beta-RollFamily: Serralysin-like metalloprotease, C-terminal domain
16 c5cxlA_
96.4
25
PDB header: toxinChain: A: PDB Molecule: bifunctional hemolysin/adenylate cyclase;PDBTitle: crystal structure of rtx domain block v of adenylate cyclase toxin2 from bordetella pertussis
17 c3p4gD_
96.4
13
PDB header: antifreeze proteinChain: D: PDB Molecule: antifreeze protein;PDBTitle: x-ray crystal structure of a hyperactive, ca2+-dependent, beta-helical2 antifreeze protein from an antarctic bacterium
18 d1sata1
96.1
21
Fold: Single-stranded right-handed beta-helixSuperfamily: beta-RollFamily: Serralysin-like metalloprotease, C-terminal domain
19 c4q1qA_
78.0
18
PDB header: cell adhesionChain: A: PDB Molecule: adhesin/invasin tiba autotransporter;PDBTitle: crystal structure of tibc-catalyzed hyper-glycosylated tiba55-3502 fragment
20 c5juhA_
46.9
22
PDB header: cell adhesionChain: A: PDB Molecule: antifreeze protein;PDBTitle: crystal structure of c-terminal domain (rv) of mpafp
21 c5ctdB_
not modelled
39.1
28
PDB header: structural proteinChain: B: PDB Molecule: collagen alpha-2(i) chain,collagen alpha-2(ix) chain;PDBTitle: crystal structure of the type ix collagen nc2 hetero-trimerization2 domain with a guest fragment a2a1a1 of type i collagen
22 c5ctdA_
not modelled
28.4
29
PDB header: structural proteinChain: A: PDB Molecule: collagen alpha-1(i) chain,collagen alpha-1(ix) chain;PDBTitle: crystal structure of the type ix collagen nc2 hetero-trimerization2 domain with a guest fragment a2a1a1 of type i collagen
23 c5ctiC_
not modelled
18.1
26
PDB header: structural proteinChain: C: PDB Molecule: collagen alpha-1(i) chain,collagen alpha-3(ix) chain;PDBTitle: crystal structure of the type ix collagen nc2 hetero-trimerization2 domain with a guest fragment a2a1a1 of type i collagen (native form)