| 1 |
|
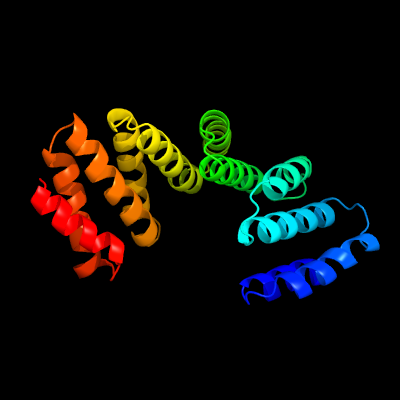
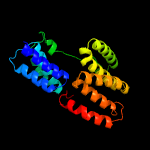
PDB 6mfv chain C
Region: 34 - 241
Aligned: 204
Modelled: 208
Confidence: 96.4%
Identity: 12%
PDB header:signaling protein
Chain: C: PDB Molecule:tetratricopeptide repeat sensor ph0952;
PDBTitle: crystal structure of the signal transduction atpase with numerous2 domains (stand) protein with a tetratricopeptide repeat sensor ph09523 from pyrococcus horikoshii
Phyre2
| 2 |
|
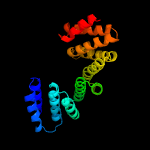
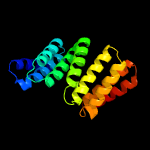
PDB 5khr chain O
Region: 57 - 241
Aligned: 183
Modelled: 183
Confidence: 90.5%
Identity: 21%
PDB header:cell cycle
Chain: O: PDB Molecule:anaphase-promoting complex subunit 5;
PDBTitle: model of human anaphase-promoting complex/cyclosome complex (apc152 deletion mutant) in complex with the e2 ube2c/ubch10 poised for3 ubiquitin ligation to substrate (apc/c-cdc20-substrate-ube2c)
Phyre2
| 3 |
|
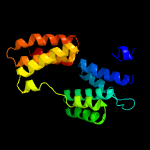
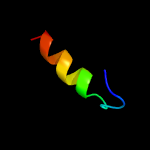
PDB 1hz4 chain A
Region: 39 - 239
Aligned: 198
Modelled: 201
Confidence: 43.7%
Identity: 17%
Fold: alpha-alpha superhelix
Superfamily: TPR-like
Family: Transcription factor MalT domain III
Phyre2
| 4 |
|
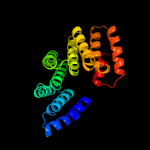
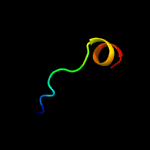
PDB 2n8i chain A
Region: 163 - 226
Aligned: 62
Modelled: 64
Confidence: 42.1%
Identity: 11%
PDB header:de novo protein
Chain: A: PDB Molecule:designed protein da05;
PDBTitle: solution nmr structure of designed protein da05, northeast structural2 genomics consortium (nesg) target or626
Phyre2
| 5 |
|
PDB 3qky chain A
Region: 31 - 93
Aligned: 63
Modelled: 63
Confidence: 32.4%
Identity: 19%
PDB header:membrane protein
Chain: A: PDB Molecule:outer membrane assembly lipoprotein yfio;
PDBTitle: crystal structure of rhodothermus marinus bamd
Phyre2
| 6 |
|
PDB 4ui9 chain O
Region: 68 - 272
Aligned: 200
Modelled: 205
Confidence: 31.8%
Identity: 18%
PDB header:cell cycle
Chain: O: PDB Molecule:anaphase-promoting complex subunit 5;
PDBTitle: atomic structure of the human anaphase-promoting complex
Phyre2
| 7 |
|
PDB 3gw4 chain B
Region: 48 - 226
Aligned: 174
Modelled: 179
Confidence: 20.9%
Identity: 20%
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of uncharacterized protein from deinococcus2 radiodurans. northeast structural genomics consortium target drr162b.
Phyre2
| 8 |
|
PDB 3hmj chain B
Region: 43 - 60
Aligned: 18
Modelled: 18
Confidence: 14.5%
Identity: 33%
PDB header:transferase
Chain: B: PDB Molecule:fatty acid synthase subunit alpha;
PDBTitle: saccharomyces cerevisiae fas type i
Phyre2
| 9 |
|
PDB 3h87 chain D
Region: 58 - 72
Aligned: 15
Modelled: 15
Confidence: 13.2%
Identity: 53%
PDB header:toxin/antitoxin
Chain: D: PDB Molecule:putative uncharacterized protein;
PDBTitle: rv0301 rv0300 toxin antitoxin complex from mycobacterium tuberculosis
Phyre2
| 10 |
|
PDB 2uv9 chain B
Region: 43 - 60
Aligned: 18
Modelled: 18
Confidence: 10.9%
Identity: 28%
PDB header:transferase
Chain: B: PDB Molecule:fatty acid synthase alpha subunits;
PDBTitle: crystal structure of fatty acid synthase from thermomyces2 lanuginosus at 3.1 angstrom resolution. this file contains3 the alpha subunits of the fatty acid synthase. the entire4 crystal structure consists of one heterododecameric fatty5 acid synthase and is described in remark 400
Phyre2
| 11 |
|
PDB 4b3y chain B
Region: 43 - 60
Aligned: 18
Modelled: 18
Confidence: 10.7%
Identity: 39%
PDB header:transferase
Chain: B: PDB Molecule:fatty acid synthase;
PDBTitle: cryo-em structure of the mycobacterial fatty acid synthase
Phyre2
| 12 |
|
PDB 2uv8 chain C
Region: 43 - 60
Aligned: 18
Modelled: 18
Confidence: 10.0%
Identity: 33%
PDB header:transferase
Chain: C: PDB Molecule:fatty acid synthase subunit alpha (fas2);
PDBTitle: crystal structure of yeast fatty acid synthase with stalled2 acyl carrier protein at 3.1 angstrom resolution
Phyre2
| 13 |
|
PDB 2vkz chain C
Region: 43 - 60
Aligned: 18
Modelled: 18
Confidence: 7.4%
Identity: 33%
PDB header:transferase
Chain: C: PDB Molecule:fatty acid synthase subunit alpha;
PDBTitle: structure of the cerulenin-inhibited fungal fatty acid synthase type i2 multienzyme complex
Phyre2
| 14 |
|
PDB 4u4c chain B
Region: 42 - 50
Aligned: 9
Modelled: 9
Confidence: 6.0%
Identity: 67%
PDB header:hydrolase
Chain: B: PDB Molecule:protein air2,poly(a) rna polymerase protein 2;
PDBTitle: the molecular architecture of the tramp complex reveals the2 organization and interplay of its two catalytic activities
Phyre2
| 15 |
|
PDB 2pff chain D
Region: 43 - 60
Aligned: 18
Modelled: 18
Confidence: 6.0%
Identity: 33%
PDB header:transferase
Chain: D: PDB Molecule:fatty acid synthase subunit alpha;
PDBTitle: structural insights of yeast fatty acid synthase
Phyre2
| 16 |
|
PDB 2pff chain G
Region: 43 - 60
Aligned: 18
Modelled: 18
Confidence: 6.0%
Identity: 33%
PDB header:transferase
Chain: G: PDB Molecule:fatty acid synthase subunit alpha;
PDBTitle: structural insights of yeast fatty acid synthase
Phyre2
| 17 |
|
PDB 2pff chain A
Region: 43 - 60
Aligned: 18
Modelled: 18
Confidence: 6.0%
Identity: 33%
PDB header:transferase
Chain: A: PDB Molecule:fatty acid synthase subunit alpha;
PDBTitle: structural insights of yeast fatty acid synthase
Phyre2
| 18 |
|
PDB 1gx5 chain A
Region: 58 - 89
Aligned: 32
Modelled: 32
Confidence: 5.9%
Identity: 31%
Fold: DNA/RNA polymerases
Superfamily: DNA/RNA polymerases
Family: RNA-dependent RNA-polymerase
Phyre2
| 19 |
|
PDB 2gbb chain A
Region: 207 - 229
Aligned: 23
Modelled: 23
Confidence: 5.3%
Identity: 17%
PDB header:isomerase
Chain: A: PDB Molecule:putative chorismate mutase;
PDBTitle: crystal structure of secreted chorismate mutase from yersinia pestis
Phyre2