1 c1kp8B_
100.0
52
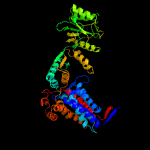
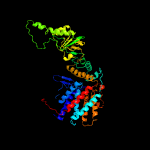
PDB header: chaperoneChain: B: PDB Molecule: groel protein;PDBTitle: structural basis for groel-assisted protein folding from the crystal2 structure of (groel-kmgatp)14 at 2.0 a resolution
2 c1we3D_
100.0
54
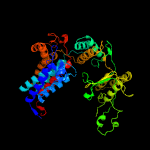
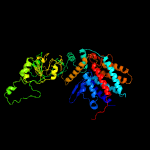
PDB header: chaperoneChain: D: PDB Molecule: cpn60(groel);PDBTitle: crystal structure of the chaperonin complex cpn60/cpn10/(adp)7 from2 thermus thermophilus
3 c4pj1E_
100.0
42
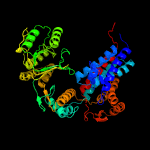
PDB header: chaperoneChain: E: PDB Molecule: 60 kda heat shock protein, mitochondrial;PDBTitle: crystal structure of the human mitochondrial chaperonin symmetrical2 'football' complex
4 c1iokE_
100.0
52
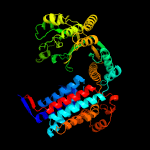
PDB header: chaperoneChain: E: PDB Molecule: chaperonin 60;PDBTitle: crystal structure of chaperonin-60 from paracoccus2 denitrificans
5 c5cdiB_
100.0
46
PDB header: chaperoneChain: B: PDB Molecule: chaperonin 60b1;PDBTitle: chloroplast chaperonin 60b1 of chlamydomonas
6 c5da8N_
100.0
50
PDB header: chaperoneChain: N: PDB Molecule: 60 kda chaperonin;PDBTitle: crystal structure of chaperonin groel from
7 c3p9ei_
100.0
19
PDB header: chaperoneChain: I: PDB Molecule: PDBTitle: the crystal structure of yeast cct reveals intrinsic asymmetry of2 eukaryotic cytosolic chaperonins
8 c3iygE_
100.0
19
PDB header: chaperoneChain: E: PDB Molecule: t-complex protein 1 subunit;PDBTitle: ca model of bovine tric/cct derived from a 4.0 angstrom cryo-em map
9 c4b2tQ_
100.0
22
PDB header: chaperoneChain: Q: PDB Molecule: t-complex protein 1 subunit theta;PDBTitle: the crystal structures of the eukaryotic chaperonin cct2 reveal its functional partitioning
10 c1q2vA_
100.0
25
PDB header: chaperoneChain: A: PDB Molecule: thermosome alpha subunit;PDBTitle: crystal structure of the chaperonin from thermococcus strain ks-12 (nucleotide-free form)
11 c3izkF_
100.0
29
PDB header: chaperoneChain: F: PDB Molecule: chaperonin;PDBTitle: mm-cpn rls deltalid with atp
12 c3iygD_
100.0
24
PDB header: chaperoneChain: D: PDB Molecule: t-complex protein 1 subunit delta;PDBTitle: ca model of bovine tric/cct derived from a 4.0 angstrom cryo-em map
13 c3ko1H_
100.0
22
PDB header: chaperoneChain: H: PDB Molecule: chaperonin;PDBTitle: cystal structure of thermosome from acidianus tengchongensis strain s5
14 c3p9en_
100.0
20
PDB header: chaperoneChain: N: PDB Molecule: PDBTitle: the crystal structure of yeast cct reveals intrinsic asymmetry of2 eukaryotic cytosolic chaperonins
15 c3iygA_
100.0
19
PDB header: chaperoneChain: A: PDB Molecule: t-complex protein 1 subunit alpha;PDBTitle: ca model of bovine tric/cct derived from a 4.0 angstrom cryo-em map
16 c4b2tE_
100.0
23
PDB header: chaperoneChain: E: PDB Molecule: t-complex protein 1 subunit epsilon;PDBTitle: the crystal structures of the eukaryotic chaperonin cct2 reveal its functional partitioning
17 c3iygZ_
100.0
20
PDB header: chaperoneChain: Z: PDB Molecule: t-complex protein 1 subunit zeta;PDBTitle: ca model of bovine tric/cct derived from a 4.0 angstrom cryo-em map
18 c4b2tD_
100.0
25
PDB header: chaperoneChain: D: PDB Molecule: t-complex protein 1 subunit delta;PDBTitle: the crystal structures of the eukaryotic chaperonin cct2 reveal its functional partitioning
19 c3p9eo_
100.0
18
PDB header: chaperoneChain: O: PDB Molecule: PDBTitle: the crystal structure of yeast cct reveals intrinsic asymmetry of2 eukaryotic cytosolic chaperonins
20 c3iygG_
100.0
18
PDB header: chaperoneChain: G: PDB Molecule: t-complex protein 1 subunit gamma;PDBTitle: ca model of bovine tric/cct derived from a 4.0 angstrom cryo-em map
21 c3losC_
not modelled
100.0
27
PDB header: chaperoneChain: C: PDB Molecule: chaperonin;PDBTitle: atomic model of mm-cpn in the closed state
22 c3p9dL_
not modelled
100.0
21
PDB header: chaperoneChain: L: PDB Molecule: t-complex protein 1 subunit delta;PDBTitle: the crystal structure of yeast cct reveals intrinsic asymmetry of2 eukaryotic cytosolic chaperonins
23 c3p9ee_
not modelled
100.0
18
PDB header: chaperoneChain: E: PDB Molecule: PDBTitle: the crystal structure of yeast cct reveals intrinsic asymmetry of2 eukaryotic cytosolic chaperonins
24 c4b2tA_
not modelled
100.0
23
PDB header: chaperoneChain: A: PDB Molecule: t-complex protein 1 subunit alpha;PDBTitle: the crystal structures of the eukaryotic chaperonin cct2 reveal its functional partitioning
25 c1a6eA_
not modelled
100.0
23
PDB header: chaperoninChain: A: PDB Molecule: thermosome (alpha subunit);PDBTitle: thermosome-mg-adp-alf3 complex
26 c3j1bA_
not modelled
100.0
22
PDB header: chaperoneChain: A: PDB Molecule: chaperonin alpha subunit;PDBTitle: cryo-em structure of 8-fold symmetric ratcpn-alpha in apo state
27 c3iygQ_
not modelled
100.0
19
PDB header: chaperoneChain: Q: PDB Molecule: t-complex protein 1 subunit theta;PDBTitle: ca model of bovine tric/cct derived from a 4.0 angstrom cryo-em map
28 c4b2tH_
not modelled
100.0
20
PDB header: chaperoneChain: H: PDB Molecule: t-complex protein 1 subunit eta;PDBTitle: the crystal structures of the eukaryotic chaperonin cct2 reveal its functional partitioning
29 c4xcgB_
not modelled
100.0
27
PDB header: chaperoneChain: B: PDB Molecule: thermosome subunit beta;PDBTitle: crystal structure of a hexadecameric tf55 complex from s.2 solfataricus, crystal form i
30 c3kttB_
not modelled
100.0
22
PDB header: chaperoneChain: B: PDB Molecule: t-complex protein 1 subunit beta;PDBTitle: atomic model of bovine tric cct2(beta) subunit derived from a 4.02 angstrom cryo-em map
31 c4b2tG_
not modelled
100.0
19
PDB header: chaperoneChain: G: PDB Molecule: t-complex protein 1 subunit gamma;PDBTitle: the crystal structures of the eukaryotic chaperonin cct2 reveal its functional partitioning
32 c3iygH_
not modelled
100.0
19
PDB header: chaperoneChain: H: PDB Molecule: t-complex protein 1 subunit eta;PDBTitle: ca model of bovine tric/cct derived from a 4.0 angstrom cryo-em map
33 c1sjpA_
not modelled
100.0
59
PDB header: chaperoneChain: A: PDB Molecule: 60 kda chaperonin 2;PDBTitle: mycobacterium tuberculosis chaperonin60.2
34 c3p9eb_
not modelled
100.0
22
PDB header: chaperoneChain: B: PDB Molecule: PDBTitle: the crystal structure of yeast cct reveals intrinsic asymmetry of2 eukaryotic cytosolic chaperonins
35 c3p9ec_
not modelled
100.0
19
PDB header: chaperoneChain: C: PDB Molecule: PDBTitle: the crystal structure of yeast cct reveals intrinsic asymmetry of2 eukaryotic cytosolic chaperonins
36 c1a6eB_
not modelled
100.0
22
PDB header: chaperoninChain: B: PDB Molecule: thermosome (beta subunit);PDBTitle: thermosome-mg-adp-alf3 complex
37 c4xciB_
not modelled
100.0
29
PDB header: chaperoneChain: B: PDB Molecule: thermosome subunit beta;PDBTitle: crystal structure of a hexadecameric tf55 complex from s.2 solfataricus, crystal form ii
38 c5x9vA_
not modelled
100.0
24
PDB header: chaperoneChain: A: PDB Molecule: thermosome, alpha subunit;PDBTitle: crystal structure of group iii chaperonin in the closed state
39 c3p9dP_
not modelled
100.0
18
PDB header: chaperoneChain: P: PDB Molecule: t-complex protein 1 subunit theta;PDBTitle: the crystal structure of yeast cct reveals intrinsic asymmetry of2 eukaryotic cytosolic chaperonins
40 c3rtkA_
not modelled
100.0
61
PDB header: chaperoneChain: A: PDB Molecule: 60 kda chaperonin 2;PDBTitle: crystal structure of cpn60.2 from mycobacterium tuberculosis at 2.8a
41 c4b2tq_
not modelled
100.0
22
PDB header: chaperoneChain: Q: PDB Molecule: t-complex protein 1 subunit theta;PDBTitle: the crystal structures of the eukaryotic chaperonin cct2 reveal its functional partitioning
42 c4b2th_
not modelled
100.0
27
PDB header: chaperoneChain: H: PDB Molecule: t-complex protein 1 subunit eta;PDBTitle: the crystal structures of the eukaryotic chaperonin cct2 reveal its functional partitioning
43 c4b2tg_
not modelled
100.0
21
PDB header: chaperoneChain: G: PDB Molecule: t-complex protein 1 subunit gamma;PDBTitle: the crystal structures of the eukaryotic chaperonin cct2 reveal its functional partitioning
44 c4b2ta_
not modelled
100.0
25
PDB header: chaperoneChain: A: PDB Molecule: t-complex protein 1 subunit alpha;PDBTitle: the crystal structures of the eukaryotic chaperonin cct2 reveal its functional partitioning
45 c4xciA_
not modelled
100.0
28
PDB header: chaperoneChain: A: PDB Molecule: thermosome subunit alpha;PDBTitle: crystal structure of a hexadecameric tf55 complex from s.2 solfataricus, crystal form ii
46 c3aq1B_
not modelled
100.0
25
PDB header: chaperoneChain: B: PDB Molecule: thermosome subunit;PDBTitle: open state monomer of a group ii chaperonin from methanococcoides2 burtonii
47 d1q3qa1
not modelled
100.0
36
Fold: GroEL equatorial domain-likeSuperfamily: GroEL equatorial domain-likeFamily: Group II chaperonin (CCT, TRIC), ATPase domain
48 d1a6db1
not modelled
100.0
34
Fold: GroEL equatorial domain-likeSuperfamily: GroEL equatorial domain-likeFamily: Group II chaperonin (CCT, TRIC), ATPase domain
49 d1a6da1
not modelled
100.0
34
Fold: GroEL equatorial domain-likeSuperfamily: GroEL equatorial domain-likeFamily: Group II chaperonin (CCT, TRIC), ATPase domain
50 d1we3a1
not modelled
100.0
59
Fold: GroEL equatorial domain-likeSuperfamily: GroEL equatorial domain-likeFamily: GroEL chaperone, ATPase domain
51 d1kp8a1
not modelled
100.0
62
Fold: GroEL equatorial domain-likeSuperfamily: GroEL equatorial domain-likeFamily: GroEL chaperone, ATPase domain
52 d1ioka1
not modelled
100.0
55
Fold: GroEL equatorial domain-likeSuperfamily: GroEL equatorial domain-likeFamily: GroEL chaperone, ATPase domain
53 d1sjpa1
not modelled
100.0
45
Fold: GroEL equatorial domain-likeSuperfamily: GroEL equatorial domain-likeFamily: GroEL chaperone, ATPase domain
54 c3m6cA_
not modelled
99.9
100
PDB header: chaperoneChain: A: PDB Molecule: 60 kda chaperonin 1;PDBTitle: crystal structure of mycobacterium tuberculosis groel1 apical domain
55 d1kida_
not modelled
99.9
53
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: GroEL apical domain-likeFamily: GroEL-like chaperone, apical domain
56 d1we3a2
not modelled
99.9
56
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: GroEL apical domain-likeFamily: GroEL-like chaperone, apical domain
57 d1sjpa2
not modelled
99.9
64
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: GroEL apical domain-likeFamily: GroEL-like chaperone, apical domain
58 c5cdjA_
not modelled
99.9
41
PDB header: chaperoneChain: A: PDB Molecule: rubisco large subunit-binding protein subunit alpha,PDBTitle: apical domain of chloroplast chaperonin 60a
59 d1oela2
not modelled
99.9
53
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: GroEL apical domain-likeFamily: GroEL-like chaperone, apical domain
60 d1dk7a_
not modelled
99.8
56
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: GroEL apical domain-likeFamily: GroEL-like chaperone, apical domain
61 d1srva_
not modelled
99.8
58
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: GroEL apical domain-likeFamily: GroEL-like chaperone, apical domain
62 d1ioka2
not modelled
99.5
59
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: GroEL apical domain-likeFamily: GroEL-like chaperone, apical domain
63 d1gmla_
not modelled
99.2
14
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: GroEL apical domain-likeFamily: Group II chaperonin (CCT, TRIC), apical domain
64 d1assa_
not modelled
98.9
16
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: GroEL apical domain-likeFamily: Group II chaperonin (CCT, TRIC), apical domain
65 d1q3qa2
not modelled
98.7
17
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: GroEL apical domain-likeFamily: Group II chaperonin (CCT, TRIC), apical domain
66 d1a6db2
not modelled
98.6
15
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: GroEL apical domain-likeFamily: Group II chaperonin (CCT, TRIC), apical domain
67 d1sjpa3
not modelled
98.4
36
Fold: GroEL-intermediate domain likeSuperfamily: GroEL-intermediate domain likeFamily: GroEL-like chaperone, intermediate domain
68 d1a6db3
not modelled
98.2
12
Fold: GroEL-intermediate domain likeSuperfamily: GroEL-intermediate domain likeFamily: Group II chaperonin (CCT, TRIC), intermediate domain
69 d1kp8a3
not modelled
98.1
35
Fold: GroEL-intermediate domain likeSuperfamily: GroEL-intermediate domain likeFamily: GroEL-like chaperone, intermediate domain
70 d1a6da3
not modelled
97.9
13
Fold: GroEL-intermediate domain likeSuperfamily: GroEL-intermediate domain likeFamily: Group II chaperonin (CCT, TRIC), intermediate domain
71 d1q3qa3
not modelled
97.9
18
Fold: GroEL-intermediate domain likeSuperfamily: GroEL-intermediate domain likeFamily: Group II chaperonin (CCT, TRIC), intermediate domain
72 d1ioka3
not modelled
96.9
49
Fold: GroEL-intermediate domain likeSuperfamily: GroEL-intermediate domain likeFamily: GroEL-like chaperone, intermediate domain
73 d1we3a3
not modelled
96.3
36
Fold: GroEL-intermediate domain likeSuperfamily: GroEL-intermediate domain likeFamily: GroEL-like chaperone, intermediate domain
74 d1vl2a2
not modelled
17.4
17
Fold: Argininosuccinate synthetase, C-terminal domainSuperfamily: Argininosuccinate synthetase, C-terminal domainFamily: Argininosuccinate synthetase, C-terminal domain
75 c3nrdB_
not modelled
13.4
8
PDB header: nucleotide binding proteinChain: B: PDB Molecule: histidine triad (hit) protein;PDBTitle: crystal structure of a histidine triad (hit) protein (smc02904) from2 sinorhizobium meliloti 1021 at 2.06 a resolution
76 d1a6qa1
not modelled
12.5
14
Fold: Another 3-helical bundleSuperfamily: Protein serine/threonine phosphatase 2C, C-terminal domainFamily: Protein serine/threonine phosphatase 2C, C-terminal domain
77 c3tekA_
not modelled
11.8
36
PDB header: dna binding proteinChain: A: PDB Molecule: thermodbp-single stranded dna binding protein;PDBTitle: thermodbp: a non-canonical single-stranded dna binding protein with a2 novel structure and mechanism
78 c4zn6B_
not modelled
11.6
23
PDB header: oxidoreductaseChain: B: PDB Molecule: 1-deoxy-d-xylulose 5-phosphate reductoisomerase;PDBTitle: x-ray crystal structure of 1-deoxy-d-xylulose 5-phosphate2 reductoisomerase (ispc) from acinetobacter baumannii
79 c5ujmB_
not modelled
10.6
9
PDB header: replicationChain: B: PDB Molecule: origin recognition complex subunit 2;PDBTitle: structure of the active form of human origin recognition complex and2 its atpase motor module
80 c4c53A_
not modelled
9.9
10
PDB header: viral proteinChain: A: PDB Molecule: pre-glycoprotein polyprotein gp complex;PDBTitle: crystal structure of guanarito virus gp2 in the post-fusion2 conformation
81 c3mkoA_
not modelled
9.8
21
PDB header: viral proteinChain: A: PDB Molecule: glycoprotein c;PDBTitle: crystal structure of the lymphocytic choriomeningitis virus membrane2 fusion glycoprotein gp2 in its postfusion conformation
82 d2hq4a1
not modelled
9.6
24
Fold: PH1570-likeSuperfamily: PH1570-likeFamily: PH1570-like
83 d1j20a2
not modelled
9.3
17
Fold: Argininosuccinate synthetase, C-terminal domainSuperfamily: Argininosuccinate synthetase, C-terminal domainFamily: Argininosuccinate synthetase, C-terminal domain
84 c5kqoA_
not modelled
9.2
27
PDB header: oxidoreductaseChain: A: PDB Molecule: 1-deoxy-d-xylulose 5-phosphate reductoisomerase;PDBTitle: 1-deoxy-d-xylulose 5-phosphate reductoisomerase from vibrio vulnificus
85 c6jqsA_
not modelled
8.8
17
PDB header: dna binding proteinChain: A: PDB Molecule: dna-binding response regulator;PDBTitle: structure of transcription factor, gere
86 c3kinB_
not modelled
8.7
19
PDB header: motor proteinChain: B: PDB Molecule: kinesin heavy chain;PDBTitle: kinesin (dimeric) from rattus norvegicus
87 d1fita_
not modelled
8.5
3
Fold: HIT-likeSuperfamily: HIT-likeFamily: HIT (HINT, histidine triad) family of protein kinase-interacting proteins
88 d1n1ea1
not modelled
8.2
12
Fold: 6-phosphogluconate dehydrogenase C-terminal domain-likeSuperfamily: 6-phosphogluconate dehydrogenase C-terminal domain-likeFamily: Glycerol-3-phosphate dehydrogenase
89 c3au9A_
not modelled
7.7
18
PDB header: isomerase/isomerase inhibitorChain: A: PDB Molecule: 1-deoxy-d-xylulose 5-phosphate reductoisomerase;PDBTitle: crystal structure of the quaternary complex-1 of an isomerase
90 c2k37A_
not modelled
7.6
11
PDB header: photosynthesisChain: A: PDB Molecule: chlorosome protein a;PDBTitle: csma
91 c4mjsQ_
not modelled
7.5
15
PDB header: transferase/protein bindingChain: Q: PDB Molecule: protein kinase c zeta type;PDBTitle: crystal structure of a pb1 complex
92 c3i24B_
not modelled
7.5
6
PDB header: hydrolaseChain: B: PDB Molecule: hit family hydrolase;PDBTitle: crystal structure of a hit family hydrolase protein from vibrio2 fischeri. northeast structural genomics consortium target id vfr176
93 c1ddiA_
not modelled
7.3
22
PDB header: oxidoreductaseChain: A: PDB Molecule: sulfite reductase [nadph] flavoprotein alpha-component;PDBTitle: crystal structure of sir-fp60
94 d1q0qa3
not modelled
7.0
24
Fold: FwdE/GAPDH domain-likeSuperfamily: Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domainFamily: Dihydrodipicolinate reductase-like
95 d1cf7b_
not modelled
6.9
22
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Cell cycle transcription factor e2f-dp
96 c4wgiA_
not modelled
6.8
11
PDB header: apoptosis/inhibitorChain: A: PDB Molecule: maltose-binding periplasmic protein,induced myeloidPDBTitle: a single diastereomer of a macrolactam core binds specifically to2 myeloid cell leukemia 1 (mcl1)
97 c2kinB_
not modelled
6.8
12
PDB header: motor proteinChain: B: PDB Molecule: kinesin;PDBTitle: kinesin (monomeric) from rattus norvegicus
98 d2caza1
not modelled
6.8
14
Fold: Long alpha-hairpinSuperfamily: Endosomal sorting complex assembly domainFamily: VPS23 C-terminal domain
99 c2cazD_
not modelled
6.7
14
PDB header: protein transportChain: D: PDB Molecule: suppressor protein stp22 of temperature-sensitive alpha-PDBTitle: escrt-i core