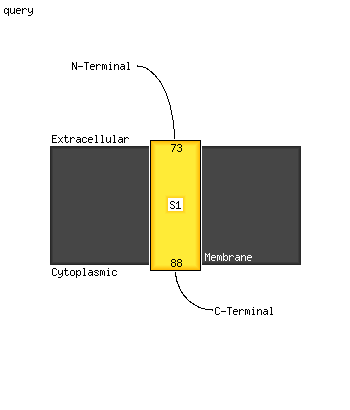
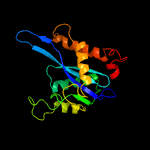
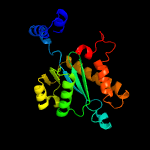
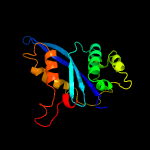
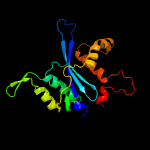
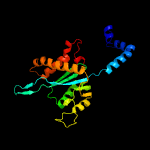
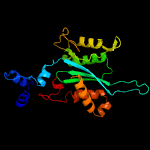
1 c4fcyA_
100.0
15
PDB header: dna binding protein/dnaChain: A: PDB Molecule: transposase;PDBTitle: crystal structure of the bacteriophage mu transpososome
2 c1bcoA_
99.9
9
PDB header: transposaseChain: A: PDB Molecule: bacteriophage mu transposase;PDBTitle: bacteriophage mu transposase core domain
3 d1bcoa2
99.8
8
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: mu transposase, core domain
4 c5u1cA_
99.8
8
PDB header: viral proteinChain: A: PDB Molecule: hiv-1 integrase, sso7d chimera;PDBTitle: structure of tetrameric hiv-1 strand transfer complex intasome
5 c5cz1B_
99.8
13
PDB header: hydrolaseChain: B: PDB Molecule: integrase;PDBTitle: crystal structure of the catalytic core domain of mmtv integrase
6 c1c0mA_
99.8
15
PDB header: transferaseChain: A: PDB Molecule: protein (integrase);PDBTitle: crystal structure of rsv two-domain integrase
7 c3f9kV_
99.8
12
PDB header: viral protein, recombinationChain: V: PDB Molecule: integrase;PDBTitle: two domain fragment of hiv-2 integrase in complex with ledgf ibd
8 c3nf9A_
99.8
10
PDB header: hydrolase/hydrolase inhibitorChain: A: PDB Molecule: integrase;PDBTitle: structural basis for a new mechanism of inhibition of hiv integrase2 identified by fragment screening and structure based design
9 c3jcaE_
99.8
15
PDB header: viral proteinChain: E: PDB Molecule: integrase;PDBTitle: core model of the mouse mammary tumor virus intasome
10 d1asua_
99.8
15
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: Retroviral integrase, catalytic domain
11 d1c0ma2
99.7
14
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: Retroviral integrase, catalytic domain
12 c5m0rF_
99.7
17
PDB header: hydrolaseChain: F: PDB Molecule: integrase;PDBTitle: cryo-em reconstruction of the maedi-visna virus (mvv) strand transfer2 complex
13 c1ex4A_
99.7
11
PDB header: viral proteinChain: A: PDB Molecule: integrase;PDBTitle: hiv-1 integrase catalytic core and c-terminal domain
14 d1exqa_
99.7
10
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: Retroviral integrase, catalytic domain
15 c3kksB_
99.7
14
PDB header: dna binding proteinChain: B: PDB Molecule: integrase;PDBTitle: crystal structure of catalytic core domain of biv integrase in crystal2 form ii
16 c4mq3A_
99.6
11
PDB header: viral proteinChain: A: PDB Molecule: integrase;PDBTitle: the 1.1 angstrom structure of catalytic core domain of fiv integrase
17 d1cxqa_
99.6
16
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: Retroviral integrase, catalytic domain
18 d1hyva_
99.6
13
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: Retroviral integrase, catalytic domain
19 c3hpgC_
99.6
17
PDB header: transferaseChain: C: PDB Molecule: integrase;PDBTitle: visna virus integrase (residues 1-219) in complex with ledgf2 ibd: examples of open integrase dimer-dimer interfaces
20 c1k6yB_
99.6
9
PDB header: transferaseChain: B: PDB Molecule: integrase;PDBTitle: crystal structure of a two-domain fragment of hiv-1 integrase
21 c5ejkG_
not modelled
99.6
15
PDB header: transferase/dnaChain: G: PDB Molecule: gag-pro-pol polyprotein;PDBTitle: crystal structure of the rous sarcoma virus intasome
22 d1c6va_
not modelled
99.5
11
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: Retroviral integrase, catalytic domain
23 c5u1cD_
not modelled
99.3
9
PDB header: viral proteinChain: D: PDB Molecule: hiv-1 integrase, sso7d chimera;PDBTitle: structure of tetrameric hiv-1 strand transfer complex intasome
24 c3l2tB_
not modelled
99.3
14
PDB header: recombination/dnaChain: B: PDB Molecule: integrase;PDBTitle: crystal structure of the prototype foamy virus (pfv) intasome in2 complex with magnesium and mk0518 (raltegravir)
25 c3dlrA_
not modelled
99.0
14
PDB header: transferaseChain: A: PDB Molecule: integrase;PDBTitle: crystal structure of the catalytic core domain from pfv integrase
26 c3l2uA_
not modelled
98.1
14
PDB header: recombination/dnaChain: A: PDB Molecule: integrase;PDBTitle: crystal structure of the prototype foamy virus (pfv)2 intasome in complex with magnesium and gs91373 (elvitegravir)
27 c3hosA_
not modelled
97.6
12
PDB header: transferase, dna binding protein/dnaChain: A: PDB Molecule: transposable element mariner, complete cds;PDBTitle: crystal structure of the mariner mos1 paired end complex with mg
28 d1bcoa1
not modelled
97.0
20
Fold: mu transposase, C-terminal domainSuperfamily: mu transposase, C-terminal domainFamily: mu transposase, C-terminal domain
29 c5cr4B_
not modelled
96.8
12
PDB header: hydrolaseChain: B: PDB Molecule: sleeping beauty transposase, sb100x;PDBTitle: crystal structure of the sleeping beauty transposase catalytic domain
30 c3f2kB_
not modelled
95.1
13
PDB header: transferaseChain: B: PDB Molecule: histone-lysine n-methyltransferase setmar;PDBTitle: structure of the transposase domain of human histone-lysine2 n-methyltransferase setmar
31 d2i5ua1
not modelled
91.7
25
Fold: DnaD domain-likeSuperfamily: DnaD domain-likeFamily: DnaD domain
32 c2zc2A_
not modelled
91.5
17
PDB header: replicationChain: A: PDB Molecule: dnad-like replication protein;PDBTitle: crystal structure of dnad-like replication protein from streptococcus2 mutans ua159, gi 24377835, residues 127-199
33 c2f7tA_
not modelled
82.0
14
PDB header: dna binding proteinChain: A: PDB Molecule: mos1 transposase;PDBTitle: crystal structure of the catalytic domain of mos1 mariner2 transposase
34 c5unkA_
not modelled
63.7
21
PDB header: dna binding proteinChain: A: PDB Molecule: sleeping beauty transposase;PDBTitle: nmr structure of the red subdomain of the sleeping beauty transposase
35 c6mzlM_
not modelled
62.8
14
PDB header: transcriptionChain: M: PDB Molecule: transcription initiation factor tfiid subunit 9, taf9;PDBTitle: human tfiid canonical state
36 d2ezia_
not modelled
48.4
25
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Recombinase DNA-binding domain
37 c2eqxA_
not modelled
45.2
20
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: kelch repeat and btb domain-containing protein 4;PDBTitle: solution structure of the back domain of kelch repeat and2 btb domain-containing protein 4
38 d2ezha_
not modelled
45.0
24
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Recombinase DNA-binding domain
39 d1mzba_
not modelled
38.9
16
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: FUR-like
40 c2o03A_
not modelled
36.1
20
PDB header: gene regulationChain: A: PDB Molecule: probable zinc uptake regulation protein furb;PDBTitle: crystal structure of furb from m. tuberculosis- a zinc uptake2 regulator
41 c4i7hA_
not modelled
34.6
16
PDB header: transcriptionChain: A: PDB Molecule: peroxide stress sensing regulator;PDBTitle: structural basis for peroxide sensing and gene regulation by perr from2 streptococcus pyogenes
42 c6dk4A_
not modelled
34.3
12
PDB header: metal transportChain: A: PDB Molecule: ferric uptake regulation protein;PDBTitle: crystal structure of campylobacter jejuni peroxide stress regulator
43 c2wbnA_
not modelled
32.6
14
PDB header: viral proteinChain: A: PDB Molecule: terminase large subunit;PDBTitle: crystal structure of the g2p (large terminase) nuclease2 domain from the bacteriophage spp1
44 c6hqaF_
not modelled
32.2
14
PDB header: transcriptionChain: F: PDB Molecule: subunit (17 kda) of tfiid and saga complexes, involved inPDBTitle: molecular structure of promoter-bound yeast tfiid
45 c2ahqA_
not modelled
30.7
19
PDB header: transcriptionChain: A: PDB Molecule: rna polymerase sigma factor rpon;PDBTitle: solution structure of the c-terminal rpon domain of sigma-2 54 from aquifex aeolicus
46 c3m5bA_
not modelled
30.5
21
PDB header: transcriptionChain: A: PDB Molecule: zinc finger and btb domain-containing protein 32;PDBTitle: crystal structure of the btb domain from fazf/zbtb32
47 c2xigA_
not modelled
29.7
28
PDB header: transcriptionChain: A: PDB Molecule: ferric uptake regulation protein;PDBTitle: the structure of the helicobacter pylori ferric uptake2 regulator fur reveals three functional metal binding sites
48 c2yy9A_
not modelled
29.3
12
PDB header: transcriptionChain: A: PDB Molecule: zinc finger and btb domain-containing protein 48;PDBTitle: crystal structure of btb domain from mouse hkr3
49 c6gfcG_
not modelled
26.9
19
PDB header: antiviral proteinChain: G: PDB Molecule: galectin-3-binding protein;PDBTitle: structure of the btb/poz domain of human 90k
50 c3mwmA_
not modelled
26.8
20
PDB header: transcriptionChain: A: PDB Molecule: putative metal uptake regulation protein;PDBTitle: graded expression of zinc-responsive genes through two regulatory2 zinc-binding sites in zur
51 c5nbcD_
not modelled
26.6
17
PDB header: dna binding proteinChain: D: PDB Molecule: ferric uptake regulation protein;PDBTitle: structure of prokaryotic transcription factors
52 c4etsB_
not modelled
25.4
8
PDB header: metal binding proteinChain: B: PDB Molecule: ferric uptake regulation protein;PDBTitle: crystal structure of campylobacter jejuni ferric uptake regulator
53 c3eyyA_
not modelled
25.1
20
PDB header: transportChain: A: PDB Molecule: putative iron uptake regulatory protein;PDBTitle: structural basis for the specialization of nur, a nickel-2 specific fur homologue, in metal sensing and dna3 recognition
54 d1stza1
not modelled
24.6
13
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Heat-inducible transcription repressor HrcA, N-terminal domain
55 c4eozC_
not modelled
24.5
20
PDB header: protein bindingChain: C: PDB Molecule: speckle-type poz protein;PDBTitle: crystal structure of the spop btb domain complexed with the cul3 n-2 terminal domain
56 c4uyiA_
not modelled
24.1
12
PDB header: hydrolaseChain: A: PDB Molecule: structure-specific endonuclease subunit slx4;PDBTitle: crystal structure of the btb domain of human slx4 (btbd12)
57 c4dkwA_
not modelled
23.0
11
PDB header: hydrolaseChain: A: PDB Molecule: large terminase protein;PDBTitle: structure of p22 large terminase nuclease domain
58 c5t9gD_
not modelled
22.9
12
PDB header: hydrolaseChain: D: PDB Molecule: glycoside hydrolase;PDBTitle: crystal structure of bugh2cwt in complex with galactoisofagomine
59 c4ypjB_
not modelled
22.5
18
PDB header: hydrolaseChain: B: PDB Molecule: beta galactosidase;PDBTitle: x-ray structure of the mutant of glycoside hydrolase
60 c6ed2A_
not modelled
22.0
16
PDB header: hydrolaseChain: A: PDB Molecule: glycosyl hydrolase family 2, tim barrel domain protein;PDBTitle: faecalibacterium prausnitzii beta-glucuronidase
61 c2fu4B_
not modelled
21.9
24
PDB header: dna binding proteinChain: B: PDB Molecule: ferric uptake regulation protein;PDBTitle: crystal structure of the dna binding domain of e.coli fur (ferric2 uptake regulator)
62 c5dmyA_
not modelled
21.9
15
PDB header: hydrolaseChain: A: PDB Molecule: beta-galactosidase;PDBTitle: beta-galactosidase - construct 33-930
63 c2w57A_
not modelled
21.9
20
PDB header: metal transportChain: A: PDB Molecule: ferric uptake regulation protein;PDBTitle: crystal structure of the vibrio cholerae ferric uptake2 regulator (fur) reveals structural rearrangement of the3 dna-binding domains
64 c3hu6B_
not modelled
20.6
20
PDB header: protein binding, ligaseChain: B: PDB Molecule: speckle-type poz protein;PDBTitle: structures of spop-substrate complexes: insights into2 molecular architectures of btb-cul3 ubiquitin ligases:3 spopmathx/btb/3-box-pucsbc1
65 c5l9wA_
not modelled
20.1
16
PDB header: ligaseChain: A: PDB Molecule: acetophenone carboxylase delta subunit;PDBTitle: crystal structure of the apc core complex
66 c6dxuB_
not modelled
20.0
16
PDB header: hydrolaseChain: B: PDB Molecule: glycosyl hydrolase family 2, tim barrel domain protein;PDBTitle: crystal structure of parabacteroides merdae beta-glucuronidase (gus)
67 d2p5ka1
not modelled
19.9
18
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Arginine repressor (ArgR), N-terminal DNA-binding domain
68 c6d1pB_
not modelled
19.6
13
PDB header: hydrolaseChain: B: PDB Molecule: glycosyl hydrolases family 2, sugar binding domain protein;PDBTitle: apo structure of bacteroides uniformis beta-glucuronidase 3
69 c5nl9B_
not modelled
19.5
16
PDB header: dna binding proteinChain: B: PDB Molecule: transcriptional regulator (fur family);PDBTitle: crystal structure of a peroxide stress regulator from leptospira2 interrogans
70 c6d0hB_
not modelled
19.2
23
PDB header: toxinChain: B: PDB Molecule: pars: cog5642 (duf2384) antitoxin;PDBTitle: part: prs adp-ribosylating toxin bound to cognate antitoxin pars
71 c4razB_
not modelled
18.7
16
PDB header: metal binding proteinChain: B: PDB Molecule: dna-binding transcriptional dual regulator of siderophorePDBTitle: crystal structure of magnetospirillum gryphiswaldense msr-1 holo-fur
72 c2mt3A_
not modelled
18.7
8
PDB header: transcriptionChain: A: PDB Molecule: rna polymerase sigma-54 factor;PDBTitle: structure of -24 dna binding domain of sigma 54 from e.coli
73 c1v85A_
not modelled
18.3
15
PDB header: apoptosisChain: A: PDB Molecule: similar to ring finger protein 36;PDBTitle: sterile alpha motif (sam) domain of mouse bifunctional2 apoptosis regulator
74 c2fe3B_
not modelled
17.7
16
PDB header: dna binding proteinChain: B: PDB Molecule: peroxide operon regulator;PDBTitle: the crystal structure of bacillus subtilis perr-zn reveals a novel2 zn(cys)4 structural redox switch
75 c2k4bA_
not modelled
17.3
11
PDB header: dna binding proteinChain: A: PDB Molecule: transcriptional regulator;PDBTitle: copr repressor structure
76 d1aoya_
not modelled
17.1
22
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Arginine repressor (ArgR), N-terminal DNA-binding domain
77 c5fd6A_
not modelled
16.9
20
PDB header: transcriptionChain: A: PDB Molecule: ferric uptake regulation protein;PDBTitle: zinc-bound manganese uptake regulator
78 c5xj2C_
not modelled
16.3
11
PDB header: transferase/rnaChain: C: PDB Molecule: uncharacterized rna methyltransferase sp_1029;PDBTitle: structure of sprlmcd with u747 rna
79 c7ceiB_
not modelled
16.1
15
PDB header: immune systemChain: B: PDB Molecule: protein (colicin e7 immunity protein);PDBTitle: the endonuclease domain of colicin e7 in complex with its inhibitor2 im7 protein
80 d1f9na1
not modelled
15.9
18
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Arginine repressor (ArgR), N-terminal DNA-binding domain
81 d1b4aa1
not modelled
15.7
21
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Arginine repressor (ArgR), N-terminal DNA-binding domain
82 c6bv7A_
not modelled
15.5
22
PDB header: membrane proteinChain: A: PDB Molecule: sodium/calcium exchanger 1;PDBTitle: nmr structure of sodium/calcium exchanger 1 (ncx1) two-helix bundle2 (thb) domain
83 c6ecaA_
not modelled
15.4
24
PDB header: hydrolaseChain: A: PDB Molecule: beta-glucuronidase;PDBTitle: lactobacillus rhamnosus beta-glucuronidase
84 d2jb0b1
not modelled
15.3
14
Fold: His-Me finger endonucleasesSuperfamily: His-Me finger endonucleasesFamily: HNH-motif
85 c6hqaK_
not modelled
15.2
24
PDB header: transcriptionChain: K: PDB Molecule: subunit (61/68 kda) of tfiid and saga complexes;PDBTitle: molecular structure of promoter-bound yeast tfiid
86 c4qkoH_
not modelled
15.2
9
PDB header: antimicrobial proteinChain: H: PDB Molecule: pyocin-s2;PDBTitle: the crystal structure of the pyocin s2 nuclease domain, immunity2 protein complex at 1.8 angstroms
87 c4uhpA_
not modelled
15.1
20
PDB header: hydrolaseChain: A: PDB Molecule: large component of pyocin ap41;PDBTitle: crystal structure of the pyocin ap41 dnase-immunity complex
88 c5ldrA_
not modelled
15.0
14
PDB header: hydrolaseChain: A: PDB Molecule: beta-d-galactosidase;PDBTitle: crystal structure of a cold-adapted dimeric beta-d-galactosidase from2 paracoccus sp. 32d strain in complex with galactose
89 c5nlbA_
not modelled
15.0
12
PDB header: ligaseChain: A: PDB Molecule: kelch-like ech-associated protein 1;PDBTitle: crystal structure of human cul3 n-terminal domain bound to keap1 btb2 and 3-box
90 c4mtdA_
not modelled
14.7
20
PDB header: dna binding protein/dnaChain: A: PDB Molecule: zinc uptake regulation protein;PDBTitle: zinc uptake regulator complexed with zinc and dna
91 c3htmB_
not modelled
14.3
29
PDB header: protein binding, ligaseChain: B: PDB Molecule: speckle-type poz protein;PDBTitle: structures of spop-substrate complexes: insights into2 molecular architectures of btb-cul3 ubiquitin ligases:3 spopbtb/3-box
92 c6mvgB_
not modelled
13.4
6
PDB header: hydrolaseChain: B: PDB Molecule: beta-glucuronidase;PDBTitle: crystal structure of fmn-binding beta-glucuronidase from ruminococcus2 gnavus
93 d1buoa_
not modelled
13.3
15
Fold: POZ domainSuperfamily: POZ domainFamily: BTB/POZ domain
94 c5t98B_
not modelled
13.2
14
PDB header: hydrolaseChain: B: PDB Molecule: glycoside hydrolase;PDBTitle: crystal structure of bugh2awt
95 c3djmA_
not modelled
12.7
16
PDB header: unknown functionChain: A: PDB Molecule: uncharacterized protein duf427;PDBTitle: crystal structure of a protein of unknown function from duf427 family2 (rsph17029_0682) from rhodobacter sphaeroides 2.4.1 at 2.51 a3 resolution
96 d2jn6a1
not modelled
12.3
22
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Cgl2762-like
97 c2o8kA_
not modelled
12.2
19
PDB header: transcription/dnaChain: A: PDB Molecule: rna polymerase sigma factor rpon;PDBTitle: nmr structure of the sigma-54 rpon domain bound to the-242 promoter element
98 c5i4rA_
not modelled
11.9
20
PDB header: toxin/antitoxinChain: A: PDB Molecule: contact-dependent inhibitor a;PDBTitle: contact-dependent inhibition system from escherichia coli nc101 -2 ternary cdia/cdii/ef-tu complex (trypsin-modified)
99 c3cmgA_
not modelled
11.8
12
PDB header: hydrolaseChain: A: PDB Molecule: putative beta-galactosidase;PDBTitle: crystal structure of putative beta-galactosidase from bacteroides2 fragilis