| 1 | c1jxaA_
|
|
 |
100.0 |
45 |
PDB header:transferase
Chain: A: PDB Molecule:glucosamine 6-phosphate synthase;
PDBTitle: glucosamine 6-phosphate synthase with glucose 6-phosphate
|
|
|
|
| 2 | c2zj3A_
|
|
 |
100.0 |
40 |
PDB header:transferase
Chain: A: PDB Molecule:glucosamine--fructose-6-phosphate
PDBTitle: isomerase domain of human glucose:fructose-6-phosphate2 amidotransferase
|
|
|
|
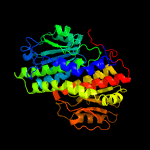
| 3 | d1moqa_
|
|
 |
100.0 |
46 |
Fold:SIS domain
Superfamily:SIS domain
Family:double-SIS domain |
|
|
|
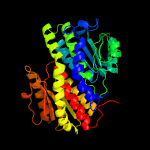
| 4 | c3tbfA_
|
|
 |
100.0 |
40 |
PDB header:transferase
Chain: A: PDB Molecule:glucosamine--fructose-6-phosphate aminotransferase
PDBTitle: c-terminal domain of glucosamine-fructose-6-phosphate aminotransferase2 from francisella tularensis.
|
|
|
|
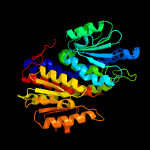
| 5 | c3odpA_
|
|
 |
100.0 |
17 |
PDB header:isomerase
Chain: A: PDB Molecule:putative tagatose-6-phosphate ketose/aldose isomerase;
PDBTitle: crystal structure of a putative tagatose-6-phosphate ketose/aldose2 isomerase (nt01cx_0292) from clostridium novyi nt at 2.35 a3 resolution
|
|
|
|
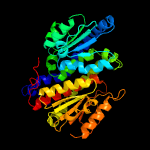
| 6 | c3i0zB_
|
|
 |
100.0 |
18 |
PDB header:isomerase
Chain: B: PDB Molecule:putative tagatose-6-phosphate ketose/aldose isomerase;
PDBTitle: crystal structure of putative putative tagatose-6-phosphate2 ketose/aldose isomerase (np_344614.1) from streptococcus pneumoniae3 tigr4 at 1.70 a resolution
|
|
|
|
| 7 | c4s1wA_
|
|
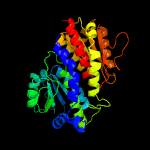 |
100.0 |
39 |
PDB header:transferase
Chain: A: PDB Molecule:glutamine--fructose-6-phosphate aminotransferase
PDBTitle: structure of a putative glutamine--fructose-6-phosphate2 aminotransferase from staphylococcus aureus subsp. aureus mu50
|
|
|
|
| 8 | c2amlB_
|
|
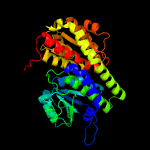 |
100.0 |
20 |
PDB header:transferase
Chain: B: PDB Molecule:sis domain protein;
PDBTitle: crystal structure of lmo0035 protein (46906266) from listeria2 monocytogenes 4b f2365 at 1.50 a resolution
|
|
|
|
| 9 | c3g68A_
|
|
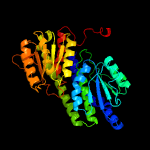 |
100.0 |
19 |
PDB header:isomerase
Chain: A: PDB Molecule:putative phosphosugar isomerase;
PDBTitle: crystal structure of a putative phosphosugar isomerase (cd3275) from2 clostridium difficile 630 at 1.80 a resolution
|
|
|
|
| 10 | c2puwA_
|
|
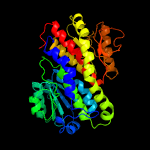 |
100.0 |
36 |
PDB header:transferase
Chain: A: PDB Molecule:isomerase domain of glutamine-fructose-6-phosphate
PDBTitle: the crystal structure of isomerase domain of glucosamine-6-phosphate2 synthase from candida albicans
|
|
|
|
| 11 | c3c3jA_
|
|
 |
100.0 |
20 |
PDB header:isomerase
Chain: A: PDB Molecule:putative tagatose-6-phosphate ketose/aldose isomerase;
PDBTitle: crystal structure of tagatose-6-phosphate ketose/aldose isomerase from2 escherichia coli
|
|
|
|
| 12 | c3fj1A_
|
|
 |
100.0 |
28 |
PDB header:isomerase
Chain: A: PDB Molecule:putative phosphosugar isomerase;
PDBTitle: crystal structure of putative phosphosugar isomerase (yp_167080.1)2 from silicibacter pomeroyi dss-3 at 1.75 a resolution
|
|
|
|
| 13 | d1j5xa_
|
|
 |
100.0 |
26 |
Fold:SIS domain
Superfamily:SIS domain
Family:double-SIS domain |
|
|
|
| 14 | c2decA_
|
|
 |
100.0 |
24 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:325aa long hypothetical protein;
PDBTitle: crystal structure of the ph0510 protein from pyrococcus horikoshii ot3
|
|
|
|
| 15 | c3fkjA_
|
|
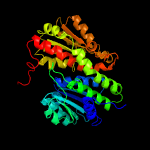 |
100.0 |
20 |
PDB header:isomerase
Chain: A: PDB Molecule:putative phosphosugar isomerases;
PDBTitle: crystal structure of a putative phosphosugar isomerase (stm_0572) from2 salmonella typhimurium lt2 at 2.12 a resolution
|
|
|
|
| 16 | c3euaD_
|
|
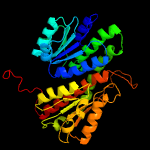 |
100.0 |
23 |
PDB header:isomerase
Chain: D: PDB Molecule:putative fructose-aminoacid-6-phosphate deglycase;
PDBTitle: crystal structure of a putative phosphosugar isomerase (bsu32610) from2 bacillus subtilis at 1.90 a resolution
|
|
|
|
| 17 | c2a3nA_
|
|
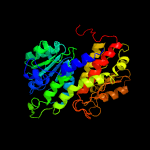 |
100.0 |
18 |
PDB header:sugar binding protein
Chain: A: PDB Molecule:putative glucosamine-fructose-6-phosphate aminotransferase;
PDBTitle: crystal structure of a putative glucosamine-fructose-6-phosphate2 aminotransferase (stm4540.s) from salmonella typhimurium lt2 at 1.353 a resolution
|
|
|
|
| 18 | c3knzA_
|
|
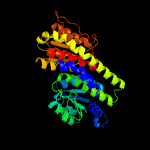 |
100.0 |
21 |
PDB header:sugar binding protein
Chain: A: PDB Molecule:putative sugar binding protein;
PDBTitle: crystal structure of putative sugar binding protein (np_459565.1) from2 salmonella typhimurium lt2 at 2.50 a resolution
|
|
|
|
| 19 | c3hbaA_
|
|
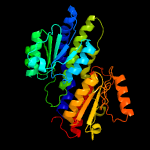 |
100.0 |
29 |
PDB header:isomerase
Chain: A: PDB Molecule:putative phosphosugar isomerase;
PDBTitle: crystal structure of a putative phosphosugar isomerase (sden_2705)2 from shewanella denitrificans os217 at 2.00 a resolution
|
|
|
|
| 20 | c1gph1_
|
|
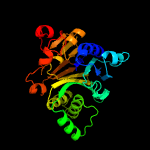 |
100.0 |
25 |
PDB header:transferase
Chain: 1: PDB Molecule:glutamine phosphoribosyl-pyrophosphate amidotransferase;
PDBTitle: structure of the allosteric regulatory enzyme of purine biosynthesis
|
|
|
|
| 21 | c1ecjB_ |
|
not modelled |
100.0 |
25 |
PDB header:transferase
Chain: B: PDB Molecule:glutamine phosphoribosylpyrophosphate
PDBTitle: escherichia coli glutamine phosphoribosylpyrophosphate2 (prpp) amidotransferase complexed with 2 amp per tetramer
|
|
|
| 22 | d1xffa_ |
|
not modelled |
100.0 |
45 |
Fold:Ntn hydrolase-like
Superfamily:N-terminal nucleophile aminohydrolases (Ntn hydrolases)
Family:Class II glutamine amidotransferases |
|
|
| 23 | d1gph12 |
|
not modelled |
100.0 |
27 |
Fold:Ntn hydrolase-like
Superfamily:N-terminal nucleophile aminohydrolases (Ntn hydrolases)
Family:Class II glutamine amidotransferases |
|
|
| 24 | d1ecfa2 |
|
not modelled |
100.0 |
27 |
Fold:Ntn hydrolase-like
Superfamily:N-terminal nucleophile aminohydrolases (Ntn hydrolases)
Family:Class II glutamine amidotransferases |
|
|
| 25 | d1x9ia_ |
|
not modelled |
100.0 |
15 |
Fold:SIS domain
Superfamily:SIS domain
Family:double-SIS domain |
|
|
| 26 | c1ct9D_ |
|
not modelled |
100.0 |
27 |
PDB header:ligase
Chain: D: PDB Molecule:asparagine synthetase b;
PDBTitle: crystal structure of asparagine synthetase b from2 escherichia coli
|
|
|
| 27 | d1ct9a2 |
|
not modelled |
100.0 |
27 |
Fold:Ntn hydrolase-like
Superfamily:N-terminal nucleophile aminohydrolases (Ntn hydrolases)
Family:Class II glutamine amidotransferases |
|
|
| 28 | d1ofda3 |
|
not modelled |
100.0 |
25 |
Fold:Ntn hydrolase-like
Superfamily:N-terminal nucleophile aminohydrolases (Ntn hydrolases)
Family:Class II glutamine amidotransferases |
|
|
| 29 | c4zfjB_ |
|
not modelled |
100.0 |
17 |
PDB header:hydrolase
Chain: B: PDB Molecule:amidohydrolase egtc;
PDBTitle: ergothioneine-biosynthetic ntn hydrolase egtc, apo form
|
|
|
| 30 | d1ea0a3 |
|
not modelled |
100.0 |
22 |
Fold:Ntn hydrolase-like
Superfamily:N-terminal nucleophile aminohydrolases (Ntn hydrolases)
Family:Class II glutamine amidotransferases |
|
|
| 31 | d1te5a_ |
|
not modelled |
100.0 |
18 |
Fold:Ntn hydrolase-like
Superfamily:N-terminal nucleophile aminohydrolases (Ntn hydrolases)
Family:Class II glutamine amidotransferases |
|
|
| 32 | c3mdnD_ |
|
not modelled |
99.9 |
26 |
PDB header:transferase
Chain: D: PDB Molecule:glutamine aminotransferase class-ii domain protein;
PDBTitle: structure of glutamine aminotransferase class-ii domain protein2 (spo2029) from silicibacter pomeroyi
|
|
|
| 33 | c4s12C_ |
|
not modelled |
99.9 |
22 |
PDB header:lyase
Chain: C: PDB Molecule:n-acetylmuramic acid 6-phosphate etherase;
PDBTitle: 1.55 angstrom crystal structure of n-acetylmuramic acid 6-phosphate2 etherase from yersinia enterocolitica.
|
|
|
| 34 | c1m1zB_ |
|
not modelled |
99.9 |
20 |
PDB header:hydrolase
Chain: B: PDB Molecule:beta-lactam synthetase;
PDBTitle: beta-lactam synthetase apo enzyme
|
|
|
| 35 | c1q15A_ |
|
not modelled |
99.9 |
25 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:cara;
PDBTitle: carbapenam synthetase
|
|
|
| 36 | d1jgta2 |
|
not modelled |
99.9 |
22 |
Fold:Ntn hydrolase-like
Superfamily:N-terminal nucleophile aminohydrolases (Ntn hydrolases)
Family:Class II glutamine amidotransferases |
|
|
| 37 | d1q15a2 |
|
not modelled |
99.9 |
23 |
Fold:Ntn hydrolase-like
Superfamily:N-terminal nucleophile aminohydrolases (Ntn hydrolases)
Family:Class II glutamine amidotransferases |
|
|
| 38 | c4lzjD_ |
|
not modelled |
99.9 |
20 |
PDB header:lyase/lyase inhibitor
Chain: D: PDB Molecule:n-acetylmuramic acid 6-phosphate etherase;
PDBTitle: crystal structure of murq from h.influenzae with bound inhibitor
|
|
|
| 39 | c2xhzC_ |
|
not modelled |
99.9 |
21 |
PDB header:isomerase
Chain: C: PDB Molecule:arabinose 5-phosphate isomerase;
PDBTitle: probing the active site of the sugar isomerase domain from e. coli2 arabinose-5-phosphate isomerase via x-ray crystallography
|
|
|
| 40 | c5uqiA_ |
|
not modelled |
99.8 |
18 |
PDB header:isomerase
Chain: A: PDB Molecule:phosphosugar isomerase;
PDBTitle: e. coli cft073 c3406 in complex with a5p
|
|
|
| 41 | c3etnD_ |
|
not modelled |
99.8 |
18 |
PDB header:isomerase
Chain: D: PDB Molecule:putative phosphosugar isomerase involved in capsule
PDBTitle: crystal structure of putative phosphosugar isomerase involved in2 capsule formation (yp_209877.1) from bacteroides fragilis nctc 93433 at 1.70 a resolution
|
|
|
| 42 | d1nria_ |
|
not modelled |
99.8 |
22 |
Fold:SIS domain
Superfamily:SIS domain
Family:mono-SIS domain |
|
|
| 43 | c1nriA_ |
|
not modelled |
99.8 |
22 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein hi0754;
PDBTitle: crystal structure of putative phosphosugar isomerase hi0754 from2 haemophilus influenzae
|
|
|
| 44 | c3fxaA_ |
|
not modelled |
99.8 |
16 |
PDB header:sugar binding protein
Chain: A: PDB Molecule:sis domain protein;
PDBTitle: crystal structure of a putative sugar-phosphate isomerase2 (lmof2365_0531) from listeria monocytogenes str. 4b f2365 at 1.60 a3 resolution
|
|
|
| 45 | c3shoA_ |
|
not modelled |
99.8 |
19 |
PDB header:transcription regulator
Chain: A: PDB Molecule:transcriptional regulator, rpir family;
PDBTitle: crystal structure of rpir transcription factor from sphaerobacter2 thermophilus (sugar isomerase domain)
|
|
|
| 46 | d1vima_ |
|
not modelled |
99.7 |
18 |
Fold:SIS domain
Superfamily:SIS domain
Family:mono-SIS domain |
|
|
| 47 | d1x94a_ |
|
not modelled |
99.7 |
14 |
Fold:SIS domain
Superfamily:SIS domain
Family:mono-SIS domain |
|
|
| 48 | d1tk9a_ |
|
not modelled |
99.7 |
16 |
Fold:SIS domain
Superfamily:SIS domain
Family:mono-SIS domain |
|
|
| 49 | c4ivnB_ |
|
not modelled |
99.7 |
13 |
PDB header:transcription regulator
Chain: B: PDB Molecule:transcriptional regulator;
PDBTitle: the vibrio vulnificus nanr protein complexed with mannac-6p
|
|
|
| 50 | d1m3sa_ |
|
not modelled |
99.7 |
18 |
Fold:SIS domain
Superfamily:SIS domain
Family:mono-SIS domain |
|
|
| 51 | c2x3yA_ |
|
not modelled |
99.6 |
15 |
PDB header:isomerase
Chain: A: PDB Molecule:phosphoheptose isomerase;
PDBTitle: crystal structure of gmha from burkholderia pseudomallei
|
|
|
| 52 | c3cvjB_ |
|
not modelled |
99.6 |
16 |
PDB header:isomerase
Chain: B: PDB Molecule:putative phosphoheptose isomerase;
PDBTitle: crystal structure of a putative phosphoheptose isomerase (bh3325) from2 bacillus halodurans c-125 at 2.00 a resolution
|
|
|
| 53 | c2vdcF_ |
|
not modelled |
99.6 |
22 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:glutamate synthase [nadph] large chain;
PDBTitle: the 9.5 a resolution structure of glutamate synthase from cryo-2 electron microscopy and its oligomerization behavior in solution:3 functional implications.
|
|
|
| 54 | d1jeoa_ |
|
not modelled |
99.6 |
17 |
Fold:SIS domain
Superfamily:SIS domain
Family:mono-SIS domain |
|
|
| 55 | c5by2A_ |
|
not modelled |
99.5 |
17 |
PDB header:isomerase
Chain: A: PDB Molecule:phosphoheptose isomerase;
PDBTitle: sedoheptulose 7-phosphate isomerase from colwellia psychrerythraea2 strain 34h
|
|
|
| 56 | d1x92a_ |
|
not modelled |
99.5 |
15 |
Fold:SIS domain
Superfamily:SIS domain
Family:mono-SIS domain |
|
|
| 57 | c3trjC_ |
|
not modelled |
99.5 |
14 |
PDB header:isomerase
Chain: C: PDB Molecule:phosphoheptose isomerase;
PDBTitle: structure of a phosphoheptose isomerase from francisella tularensis
|
|
|
| 58 | c1lm1A_ |
|
not modelled |
99.5 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:ferredoxin-dependent glutamate synthase;
PDBTitle: structural studies on the synchronization of catalytic centers in2 glutamate synthase: native enzyme
|
|
|
| 59 | c5i01B_ |
|
not modelled |
99.4 |
16 |
PDB header:isomerase
Chain: B: PDB Molecule:phosphoheptose isomerase;
PDBTitle: structure of phosphoheptose isomerase gmha from neisseria gonorrhoeae
|
|
|
| 60 | c2yvaB_ |
|
not modelled |
99.4 |
16 |
PDB header:dna binding protein
Chain: B: PDB Molecule:dnaa initiator-associating protein diaa;
PDBTitle: crystal structure of escherichia coli diaa
|
|
|
| 61 | c4bbaA_ |
|
not modelled |
99.3 |
12 |
PDB header:protein-binding protein
Chain: A: PDB Molecule:glucokinase regulatory protein;
PDBTitle: crystal structure of glucokinase regulatory protein complexed to2 phosphate
|
|
|
| 62 | c4lc9A_ |
|
not modelled |
99.3 |
13 |
PDB header:transferase/transferase regulator
Chain: A: PDB Molecule:glucokinase regulatory protein;
PDBTitle: structural basis for regulation of human glucokinase by glucokinase2 regulatory protein
|
|
|
| 63 | c3w0lD_ |
|
not modelled |
99.2 |
15 |
PDB header:transferase/transferase inhibitor
Chain: D: PDB Molecule:glucokinase regulatory protein;
PDBTitle: the crystal structure of xenopus glucokinase and glucokinase2 regulatory protein complex
|
|
|
| 64 | d1wiwa_ |
|
not modelled |
99.0 |
15 |
Fold:SIS domain
Superfamily:SIS domain
Family:double-SIS domain |
|
|
| 65 | c2q8nB_ |
|
not modelled |
98.4 |
19 |
PDB header:isomerase
Chain: B: PDB Molecule:glucose-6-phosphate isomerase;
PDBTitle: crystal structure of glucose-6-phosphate isomerase (ec 5.3.1.9)2 (tm1385) from thermotoga maritima at 1.82 a resolution
|
|
|
| 66 | d1c7qa_ |
|
not modelled |
98.1 |
17 |
Fold:SIS domain
Superfamily:SIS domain
Family:Phosphoglucose isomerase, PGI |
|
|
| 67 | c1zzgB_ |
|
not modelled |
98.1 |
18 |
PDB header:isomerase
Chain: B: PDB Molecule:glucose-6-phosphate isomerase;
PDBTitle: crystal structure of hypothetical protein tt0462 from thermus2 thermophilus hb8
|
|
|
| 68 | c6otuA_ |
|
not modelled |
98.1 |
10 |
PDB header:isomerase
Chain: A: PDB Molecule:glucose-6-phosphate isomerase;
PDBTitle: crystal structure of a glucose-6-phosphate isomerase from chlamydia2 trachomatis d/uw-3/cx
|
|
|
| 69 | c3ff1B_ |
|
not modelled |
98.1 |
23 |
PDB header:isomerase
Chain: B: PDB Molecule:glucose-6-phosphate isomerase;
PDBTitle: structure of glucose 6-phosphate isomerase from staphylococcus aureus
|
|
|
| 70 | d1gzda_ |
|
not modelled |
98.0 |
17 |
Fold:SIS domain
Superfamily:SIS domain
Family:Phosphoglucose isomerase, PGI |
|
|
| 71 | c3hjbA_ |
|
not modelled |
98.0 |
14 |
PDB header:isomerase
Chain: A: PDB Molecule:glucose-6-phosphate isomerase;
PDBTitle: 1.5 angstrom crystal structure of glucose-6-phosphate isomerase from2 vibrio cholerae.
|
|
|
| 72 | d1q50a_ |
|
not modelled |
98.0 |
14 |
Fold:SIS domain
Superfamily:SIS domain
Family:Phosphoglucose isomerase, PGI |
|
|
| 73 | c4em6D_ |
|
not modelled |
98.0 |
17 |
PDB header:isomerase
Chain: D: PDB Molecule:glucose-6-phosphate isomerase;
PDBTitle: the structure of glucose-6-phosphate isomerase (gpi) from brucella2 melitensis
|
|
|
| 74 | c2wu8A_ |
|
not modelled |
98.0 |
14 |
PDB header:isomerase
Chain: A: PDB Molecule:glucose-6-phosphate isomerase;
PDBTitle: structural studies of phosphoglucose isomerase from2 mycobacterium tuberculosis h37rv
|
|
|
| 75 | c3ljkA_ |
|
not modelled |
97.9 |
18 |
PDB header:isomerase
Chain: A: PDB Molecule:glucose-6-phosphate isomerase;
PDBTitle: glucose-6-phosphate isomerase from francisella tularensis.
|
|
|
| 76 | d1iata_ |
|
not modelled |
97.9 |
16 |
Fold:SIS domain
Superfamily:SIS domain
Family:Phosphoglucose isomerase, PGI |
|
|
| 77 | c1t10A_ |
|
not modelled |
97.9 |
14 |
PDB header:isomerase
Chain: A: PDB Molecule:glucose-6-phosphate isomerase;
PDBTitle: phosphoglucose isomerase from leishmania mexicana in complex with2 substrate d-fructose-6-phosphate
|
|
|
| 78 | d1hm5a_ |
|
not modelled |
97.9 |
15 |
Fold:SIS domain
Superfamily:SIS domain
Family:Phosphoglucose isomerase, PGI |
|
|
| 79 | d1u0fa_ |
|
not modelled |
97.9 |
15 |
Fold:SIS domain
Superfamily:SIS domain
Family:Phosphoglucose isomerase, PGI |
|
|
| 80 | c3ujhB_ |
|
not modelled |
97.8 |
18 |
PDB header:isomerase
Chain: B: PDB Molecule:glucose-6-phosphate isomerase;
PDBTitle: crystal structure of substrate-bound glucose-6-phosphate isomerase2 from toxoplasma gondii
|
|
|
| 81 | c6bzcA_ |
|
not modelled |
97.8 |
15 |
PDB header:isomerase
Chain: A: PDB Molecule:glucose-6-phosphate isomerase;
PDBTitle: crystal structure of glucose-6-phosphate isomerase from2 elizabethkingia anophelis with bound glucose-6-phosphate
|
|
|
| 82 | c3pr3B_ |
|
not modelled |
97.8 |
17 |
PDB header:isomerase
Chain: B: PDB Molecule:glucose-6-phosphate isomerase;
PDBTitle: crystal structure of plasmodium falciparum glucose-6-phosphate2 isomerase (pf14_0341) in complex with fructose-6-phosphate
|
|
|
| 83 | c2o2cB_ |
|
not modelled |
97.8 |
15 |
PDB header:isomerase
Chain: B: PDB Molecule:glucose-6-phosphate isomerase, glycosomal;
PDBTitle: crystal structure of phosphoglucose isomerase from t. brucei2 containing glucose-6-phosphate in the active site
|
|
|
| 84 | c3nbuC_ |
|
not modelled |
97.7 |
15 |
PDB header:isomerase
Chain: C: PDB Molecule:glucose-6-phosphate isomerase;
PDBTitle: crystal structure of pgi glucosephosphate isomerase
|
|
|
| 85 | c4qfhA_ |
|
not modelled |
97.7 |
15 |
PDB header:isomerase
Chain: A: PDB Molecule:glucose-6-phosphate isomerase;
PDBTitle: structure of a glucose-6-phosphate isomerase from trypanosoma cruzi
|
|
|
| 86 | d2iv2x2 |
|
not modelled |
94.8 |
17 |
Fold:Formate dehydrogenase/DMSO reductase, domains 1-3
Superfamily:Formate dehydrogenase/DMSO reductase, domains 1-3
Family:Formate dehydrogenase/DMSO reductase, domains 1-3 |
|
|
| 87 | d1tmoa2 |
|
not modelled |
94.6 |
14 |
Fold:Formate dehydrogenase/DMSO reductase, domains 1-3
Superfamily:Formate dehydrogenase/DMSO reductase, domains 1-3
Family:Formate dehydrogenase/DMSO reductase, domains 1-3 |
|
|
| 88 | c1tmoA_ |
|
not modelled |
94.1 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:trimethylamine n-oxide reductase;
PDBTitle: trimethylamine n-oxide reductase from shewanella massilia
|
|
|
| 89 | c2iv2X_ |
|
not modelled |
93.9 |
16 |
PDB header:oxidoreductase
Chain: X: PDB Molecule:formate dehydrogenase h;
PDBTitle: reinterpretation of reduced form of formate dehydrogenase h from e.2 coli
|
|
|
| 90 | c2ivfA_ |
|
not modelled |
93.0 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:ethylbenzene dehydrogenase alpha-subunit;
PDBTitle: ethylbenzene dehydrogenase from aromatoleum aromaticum
|
|
|
| 91 | c2e7zA_ |
|
not modelled |
91.8 |
15 |
PDB header:lyase
Chain: A: PDB Molecule:acetylene hydratase ahy;
PDBTitle: acetylene hydratase from pelobacter acetylenicus
|
|
|
| 92 | d1dmra2 |
|
not modelled |
90.1 |
14 |
Fold:Formate dehydrogenase/DMSO reductase, domains 1-3
Superfamily:Formate dehydrogenase/DMSO reductase, domains 1-3
Family:Formate dehydrogenase/DMSO reductase, domains 1-3 |
|
|
| 93 | d2jioa2 |
|
not modelled |
90.0 |
17 |
Fold:Formate dehydrogenase/DMSO reductase, domains 1-3
Superfamily:Formate dehydrogenase/DMSO reductase, domains 1-3
Family:Formate dehydrogenase/DMSO reductase, domains 1-3 |
|
|
| 94 | d1eu1a2 |
|
not modelled |
89.9 |
16 |
Fold:Formate dehydrogenase/DMSO reductase, domains 1-3
Superfamily:Formate dehydrogenase/DMSO reductase, domains 1-3
Family:Formate dehydrogenase/DMSO reductase, domains 1-3 |
|
|
| 95 | c3fpjA_ |
|
not modelled |
89.5 |
11 |
PDB header:biosynthetic protein, transferase
Chain: A: PDB Molecule:putative uncharacterized protein;
PDBTitle: crystal structure of e81q mutant of mtnas in complex with s-2 adenosylmethionine
|
|
|
| 96 | c3iz6A_ |
|
not modelled |
89.1 |
14 |
PDB header:ribosome
Chain: A: PDB Molecule:40s ribosomal protein sa (s2p);
PDBTitle: localization of the small subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
|
|
|
| 97 | c1h5nC_ |
|
not modelled |
88.7 |
14 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:dmso reductase;
PDBTitle: dmso reductase modified by the presence of dms and air
|
|
|
| 98 | c1vlfQ_ |
|
not modelled |
88.6 |
13 |
PDB header:oxidoreductase
Chain: Q: PDB Molecule:pyrogallol hydroxytransferase large subunit;
PDBTitle: crystal structure of pyrogallol-phloroglucinol transhydroxylase from2 pelobacter acidigallici complexed with inhibitor 1,2,4,5-3 tetrahydroxy-benzene
|
|
|
| 99 | d1p3da1 |
|
not modelled |
88.4 |
17 |
Fold:MurCD N-terminal domain
Superfamily:MurCD N-terminal domain
Family:MurCD N-terminal domain |
|
|
| 100 | c5ucgE_ |
|
not modelled |
87.7 |
15 |
PDB header:hydrolase
Chain: E: PDB Molecule:stage ii sporulation protein e;
PDBTitle: structure of the pp2c phosphatase domain and a fragment of the2 regulatory domain of the cell fate determinant spoiie from bacillus3 subtilis
|
|
|
| 101 | c2f00A_ |
|
not modelled |
84.7 |
19 |
PDB header:ligase
Chain: A: PDB Molecule:udp-n-acetylmuramate--l-alanine ligase;
PDBTitle: escherichia coli murc
|
|
|
| 102 | d1vlfm2 |
|
not modelled |
83.6 |
16 |
Fold:Formate dehydrogenase/DMSO reductase, domains 1-3
Superfamily:Formate dehydrogenase/DMSO reductase, domains 1-3
Family:Formate dehydrogenase/DMSO reductase, domains 1-3 |
|
|
| 103 | c2vpyE_ |
|
not modelled |
82.8 |
15 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:thiosulfate reductase;
PDBTitle: polysulfide reductase with bound quinone inhibitor,2 pentachlorophenol (pcp)
|
|
|
| 104 | c1eu1A_ |
|
not modelled |
80.5 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dimethyl sulfoxide reductase;
PDBTitle: the crystal structure of rhodobacter sphaeroides dimethylsulfoxide2 reductase reveals two distinct molybdenum coordination environments.
|
|
|
| 105 | c1h0hA_ |
|
not modelled |
78.1 |
18 |
PDB header:electron transport
Chain: A: PDB Molecule:formate dehydrogenase subunit alpha;
PDBTitle: tungsten containing formate dehydrogenase from desulfovibrio gigas
|
|
|
| 106 | c4ffnA_ |
|
not modelled |
77.7 |
19 |
PDB header:ligase/substrate
Chain: A: PDB Molecule:pylc;
PDBTitle: pylc in complex with d-ornithine and amppnp
|
|
|
| 107 | c6cz7C_ |
|
not modelled |
77.5 |
11 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:arra;
PDBTitle: the arsenate respiratory reductase (arr) complex from shewanella sp.2 ana-3
|
|
|
| 108 | c2e4gB_ |
|
not modelled |
77.0 |
27 |
PDB header:biosynthetic protein, flavoprotein
Chain: B: PDB Molecule:tryptophan halogenase;
PDBTitle: rebh with bound l-trp
|
|
|
| 109 | c6ib5B_ |
|
not modelled |
76.8 |
21 |
PDB header:flavoprotein
Chain: B: PDB Molecule:tryptophan 6-halogenase;
PDBTitle: mutant of flavin-dependent tryptophan halogenase thal with altered2 regioselectivity (thal-rebh5)
|
|
|
| 110 | c3zt9A_ |
|
not modelled |
73.0 |
11 |
PDB header:hydrolase
Chain: A: PDB Molecule:serine phosphatase;
PDBTitle: the bacterial stressosome: a modular system that has been adapted to2 control secondary messenger signaling
|
|
|
| 111 | c3jx9B_ |
|
not modelled |
71.3 |
14 |
PDB header:isomerase
Chain: B: PDB Molecule:putative phosphoheptose isomerase;
PDBTitle: crystal structure of putative phosphoheptose isomerase2 (yp_001815198.1) from exiguobacterium sp. 255-15 at 1.95 a resolution
|
|
|
| 112 | c5fn0C_ |
|
not modelled |
71.0 |
14 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:kynurenine 3-monooxygenase;
PDBTitle: crystal structure of pseudomonas fluorescens kynurenine-3-2 monooxygenase (kmo) in complex with gsk180
|
|
|
| 113 | c2xznB_ |
|
not modelled |
70.8 |
14 |
PDB header:ribosome
Chain: B: PDB Molecule:rps0e;
PDBTitle: crystal structure of the eukaryotic 40s ribosomal2 subunit in complex with initiation factor 1. this file3 contains the 40s subunit and initiation factor for4 molecule 2
|
|
|
| 114 | d1g8ka2 |
|
not modelled |
69.1 |
11 |
Fold:Formate dehydrogenase/DMSO reductase, domains 1-3
Superfamily:Formate dehydrogenase/DMSO reductase, domains 1-3
Family:Formate dehydrogenase/DMSO reductase, domains 1-3 |
|
|
| 115 | d1h0ha2 |
|
not modelled |
68.9 |
19 |
Fold:Formate dehydrogenase/DMSO reductase, domains 1-3
Superfamily:Formate dehydrogenase/DMSO reductase, domains 1-3
Family:Formate dehydrogenase/DMSO reductase, domains 1-3 |
|
|
| 116 | c2pyxA_ |
|
not modelled |
68.8 |
18 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:tryptophan halogenase;
PDBTitle: crystal structure of tryptophan halogenase (yp_750003.1) from2 shewanella frigidimarina ncimb 400 at 1.50 a resolution
|
|
|
| 117 | c3x0vA_ |
|
not modelled |
68.5 |
26 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:l-lysine oxidase;
PDBTitle: structure of l-lysine oxidase
|
|
|
| 118 | c6frlA_ |
|
not modelled |
68.5 |
30 |
PDB header:flavoprotein
Chain: A: PDB Molecule:tryptophan halogenase superfamily;
PDBTitle: brvh, a flavin-dependent halogenase from brevundimonas sp. bal3
|
|
|
| 119 | d1j6ua1 |
|
not modelled |
68.5 |
16 |
Fold:MurCD N-terminal domain
Superfamily:MurCD N-terminal domain
Family:MurCD N-terminal domain |
|
|
| 120 | d1yc5a1 |
|
not modelled |
67.7 |
9 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Sir2 family of transcriptional regulators |
|
|