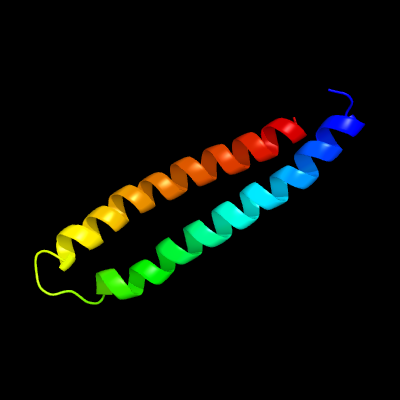
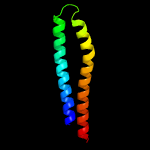
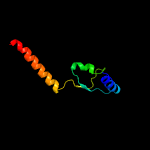
1 d1wa8a1
94.6
19
Fold: Ferritin-likeSuperfamily: EsxAB dimer-likeFamily: ESAT-6 like
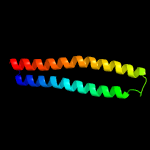
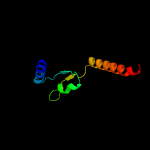
2 c4lwsA_
94.3
16
PDB header: unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: esxa : esxb (semet) hetero-dimer from thermomonospora curvata
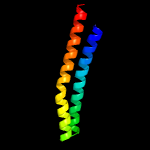
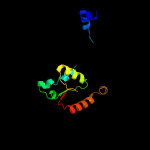
3 d1wa8b1
94.3
18
Fold: Ferritin-likeSuperfamily: EsxAB dimer-likeFamily: ESAT-6 like
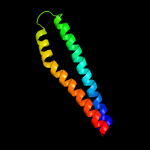
4 c4lwsB_
94.2
14
PDB header: unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: esxa : esxb (semet) hetero-dimer from thermomonospora curvata
5 c2vs0B_
93.0
14
PDB header: cell invasionChain: B: PDB Molecule: virulence factor esxa;PDBTitle: structural analysis of homodimeric staphylococcal aureus2 virulence factor esxa
6 c4iogD_
91.9
9
PDB header: unknown functionChain: D: PDB Molecule: secreted protein esxb;PDBTitle: the crystal structure of a secreted protein esxb (wild-type, in p212 space group) from bacillus anthracis str. sterne
7 c3zbhC_
91.3
15
PDB header: unknown functionChain: C: PDB Molecule: esxa;PDBTitle: geobacillus thermodenitrificans esxa crystal form i
8 c3gvmA_
91.2
17
PDB header: viral proteinChain: A: PDB Molecule: putative uncharacterized protein sag1039;PDBTitle: structure of the homodimeric wxg-100 family protein from streptococcus2 agalactiae
9 c2kg7B_
88.8
15
PDB header: unknown functionChain: B: PDB Molecule: esat-6-like protein esxh;PDBTitle: structure and features of the complex formed by the tuberculosis2 virulence factors rv0287 and rv0288
10 d2g38b1
74.1
10
Fold: Ferritin-likeSuperfamily: PE/PPE dimer-likeFamily: PPE
11 c2g38B_
74.1
10
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: ppe family protein;PDBTitle: a pe/ppe protein complex from mycobacterium tuberculosis
12 c2i56A_
70.3
20
PDB header: isomerase, metal-binding proteinChain: A: PDB Molecule: l-rhamnose isomerase;PDBTitle: crystal structure of l-rhamnose isomerase from pseudomonas2 stutzeri with l-rhamnose
13 c5hypB_
61.3
24
PDB header: immune systemChain: B: PDB Molecule: m28 protein;PDBTitle: structure of human c4b-binding protein alpha cain ccp domains 1 and 22 in complex with the hypervariable region of group a streptococcus m283 protein
14 c6ogdB_
60.9
12
PDB header: toxinChain: B: PDB Molecule: toxin subunit yena2;PDBTitle: cryo-em structure of yentca in its prepore state
15 c3d3kD_
56.6
13
PDB header: protein bindingChain: D: PDB Molecule: enhancer of mrna-decapping protein 3;PDBTitle: crystal structure of human edc3p
16 c4i0xJ_
54.9
17
PDB header: structural genomics, unknown functionChain: J: PDB Molecule: esat-6-like protein mab_3113;PDBTitle: crystal structure of the mycobacterum abscessus esxef (mab_3112-2 mab_3113) complex
17 d1izca_
53.5
16
Fold: TIM beta/alpha-barrelSuperfamily: Phosphoenolpyruvate/pyruvate domainFamily: HpcH/HpaI aldolase
18 c1izcA_
53.5
16
PDB header: lyaseChain: A: PDB Molecule: macrophomate synthase intermolecular diels-alderase;PDBTitle: crystal structure analysis of macrophomate synthase
19 d1qh8b_
49.6
21
Fold: Chelatase-likeSuperfamily: "Helical backbone" metal receptorFamily: Nitrogenase iron-molybdenum protein
20 d1nt2b_
48.2
28
Fold: Nop domainSuperfamily: Nop domainFamily: Nop domain
21 c4ffcD_
not modelled
41.7
23
PDB header: transferaseChain: D: PDB Molecule: 4-aminobutyrate aminotransferase (gabt);PDBTitle: crystal structure of a 4-aminobutyrate aminotransferase (gabt) from2 mycobacterium abscessus
22 c4i0xA_
not modelled
40.3
20
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: esat-6-like protein mab_3112;PDBTitle: crystal structure of the mycobacterum abscessus esxef (mab_3112-2 mab_3113) complex
23 c2ykyB_
not modelled
37.0
23
PDB header: transferaseChain: B: PDB Molecule: beta-transaminase;PDBTitle: structural determinants of the beta-selectivity of a bacterial2 aminotransferase
24 c3e04C_
not modelled
36.4
20
PDB header: lyaseChain: C: PDB Molecule: fumarate hydratase;PDBTitle: crystal structure of human fumarate hydratase
25 c5ghfB_
not modelled
35.9
19
PDB header: transferaseChain: B: PDB Molecule: aminotransferase class-iii;PDBTitle: transaminase with l-ala
26 d1dxea_
not modelled
35.8
18
Fold: TIM beta/alpha-barrelSuperfamily: Phosphoenolpyruvate/pyruvate domainFamily: HpcH/HpaI aldolase
27 c3m1eA_
not modelled
35.0
14
PDB header: transcription regulatorChain: A: PDB Molecule: hth-type transcriptional regulator benm;PDBTitle: crystal structure of benm_dbd
28 c2uvaI_
not modelled
34.8
21
PDB header: transferaseChain: I: PDB Molecule: fatty acid synthase beta subunits;PDBTitle: crystal structure of fatty acid synthase from thermomyces2 lanuginosus at 3.1 angstrom resolution. this file contains3 the beta subunits of the fatty acid synthase. the entire4 crystal structure consists of one heterododecameric fatty5 acid synthase and is described in remark 400
29 c4gioA_
not modelled
33.8
42
PDB header: unknown functionChain: A: PDB Molecule: putative lipoprotein;PDBTitle: crystal structure of campylobacter jejuni cj0090
30 c3w3fB_
not modelled
33.6
17
PDB header: biosynthetic proteinChain: B: PDB Molecule: uncharacterized protein blr2150;PDBTitle: crystal structure of ent-kaurene synthase bjks from bradyrhizobium2 japonicum
31 c3id6A_
not modelled
33.5
9
PDB header: transferaseChain: A: PDB Molecule: pre mrna splicing protein;PDBTitle: crystal structure of sulfolobus solfataricus nop5 (1-262) and2 fibrillarin complex
32 c6akmA_
not modelled
33.2
19
PDB header: protein bindingChain: A: PDB Molecule: suppressor of ikbke 1;PDBTitle: crystal structure of slmap-sike1 complex
33 d1xm5a_
not modelled
32.7
39
Fold: Zincin-likeSuperfamily: Metalloproteases ("zincins"), catalytic domainFamily: Predicted metal-dependent hydrolase
34 c2yy0D_
not modelled
30.1
16
PDB header: transcriptionChain: D: PDB Molecule: c-myc-binding protein;PDBTitle: crystal structure of ms0802, c-myc-1 binding protein domain2 from homo sapiens
35 d1qm4a2
not modelled
29.6
19
Fold: S-adenosylmethionine synthetaseSuperfamily: S-adenosylmethionine synthetaseFamily: S-adenosylmethionine synthetase
36 c3n8uB_
not modelled
29.0
16
PDB header: hydrolaseChain: B: PDB Molecule: imelysin peptidase;PDBTitle: crystal structure of an imelysin peptidase (bacova_03801) from2 bacteroides ovatus at 1.44 a resolution
37 c3oksB_
not modelled
28.1
15
PDB header: transferaseChain: B: PDB Molecule: 4-aminobutyrate transaminase;PDBTitle: crystal structure of 4-aminobutyrate transaminase from mycobacterium2 smegmatis
38 d1sffa_
not modelled
27.6
19
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: GABA-aminotransferase-like
39 d2d0ta1
not modelled
27.1
13
Fold: Indolic compounds 2,3-dioxygenase-likeSuperfamily: Indolic compounds 2,3-dioxygenase-likeFamily: Indoleamine 2,3-dioxygenase-like
40 c3n5mD_
not modelled
26.7
16
PDB header: transferaseChain: D: PDB Molecule: adenosylmethionine-8-amino-7-oxononanoate aminotransferase;PDBTitle: crystals structure of a bacillus anthracis aminotransferase
41 c2n39A_
not modelled
26.0
17
PDB header: dna binding proteinChain: A: PDB Molecule: chromodomain-helicase-dna-binding protein 1;PDBTitle: nmr solution structure of a c-terminal domain of the chromodomain2 helicase dna-binding protein 1
42 d2fr2a1
not modelled
25.4
33
Fold: LipocalinsSuperfamily: LipocalinsFamily: Rv2717c-like
43 d1zoda1
not modelled
25.3
16
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: GABA-aminotransferase-like
44 c1wqsA_
not modelled
25.0
55
PDB header: hydrolaseChain: A: PDB Molecule: 3c-like protease;PDBTitle: crystal structure of norovirus 3c-like protease
45 d1c3ca_
not modelled
24.7
12
Fold: L-aspartase-likeSuperfamily: L-aspartase-likeFamily: L-aspartase/fumarase
46 c3hhgF_
not modelled
24.6
16
PDB header: transcription regulatorChain: F: PDB Molecule: transcriptional regulator, lysr family;PDBTitle: structure of crga, a lysr-type transcriptional regulator from2 neisseria meningitidis.
47 c5fo5A_
not modelled
24.1
11
PDB header: transcriptionChain: A: PDB Molecule: hth-type transcriptional regulator metr;PDBTitle: structure of the dna-binding domain of escherichia coli methionine2 biosynthesis regulator metr
48 d1ob0a2
not modelled
24.0
36
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain
49 d1tvia_
not modelled
22.5
29
Fold: Zincin-likeSuperfamily: Metalloproteases ("zincins"), catalytic domainFamily: Predicted metal-dependent hydrolase
50 d1u94a2
not modelled
22.4
25
Fold: Anti-LPS factor/recA domainSuperfamily: RecA protein, C-terminal domainFamily: RecA protein, C-terminal domain
51 c3o0lB_
not modelled
21.8
25
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a pfam duf1425 family member (shew_1734) from2 shewanella sp. pv-4 at 1.81 a resolution
52 c4arpB_
not modelled
21.5
16
PDB header: hydrolaseChain: B: PDB Molecule: pesticin;PDBTitle: structure of the inactive pesticin e178a mutant
53 d1xp8a2
not modelled
21.1
21
Fold: Anti-LPS factor/recA domainSuperfamily: RecA protein, C-terminal domainFamily: RecA protein, C-terminal domain
54 c2v5jB_
not modelled
21.0
18
PDB header: lyaseChain: B: PDB Molecule: 2,4-dihydroxyhept-2-ene-1,7-dioic acid aldolase;PDBTitle: apo class ii aldolase hpch
55 d2al6a3
not modelled
20.5
15
Fold: beta-Grasp (ubiquitin-like)Superfamily: Ubiquitin-likeFamily: First domain of FERM
56 d1e43a2
not modelled
20.5
28
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain
57 c3id5E_
not modelled
20.4
10
PDB header: transferase/ribosomal protein/rnaChain: E: PDB Molecule: pre mrna splicing protein;PDBTitle: crystal structure of sulfolobus solfataricus c/d rnp assembled with2 nop5, fibrillarin, l7ae and a split half c/d rna
58 c1bplA_
not modelled
20.3
32
PDB header: glycosyltransferaseChain: A: PDB Molecule: alpha-1,4-glucan-4-glucanohydrolase;PDBTitle: glycosyltransferase
59 c4gq6A_
not modelled
20.2
33
PDB header: transcription/transcription inhibitorChain: A: PDB Molecule: menin;PDBTitle: human menin in complex with mll peptide
60 c2fwvA_
not modelled
20.1
33
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein mtubf_01000852;PDBTitle: crystal structure of rv0813
61 c2jq3A_
not modelled
19.9
27
PDB header: lipid binding proteinChain: A: PDB Molecule: apolipoprotein c-iii;PDBTitle: structure and dynamics of human apolipoprotein c-iii
62 c2nnwC_
not modelled
19.7
14
PDB header: transferaseChain: C: PDB Molecule: nop5/nop56 related protein;PDBTitle: alternative conformations of nop56/58-fibrillarin complex and2 implication for induced-fit assenly of box c/d rnps
63 d1jm6a1
not modelled
19.5
23
Fold: Bromodomain-likeSuperfamily: alpha-ketoacid dehydrogenase kinase, N-terminal domainFamily: alpha-ketoacid dehydrogenase kinase, N-terminal domain
64 c3re2A_
not modelled
19.5
33
PDB header: unknown functionChain: A: PDB Molecule: predicted protein;PDBTitle: crystal structure of menin reveals the binding site for mixed lineage2 leukemia (mll) protein
65 d2c0sa1
not modelled
19.4
23
Fold: ROP-likeSuperfamily: BAS1536-likeFamily: BAS1536-like
66 c4mf4F_
not modelled
19.2
20
PDB header: lyaseChain: F: PDB Molecule: hpch/hpai aldolase/citrate lyase family protein;PDBTitle: crystal structure of a hpch/hpal aldolase/citrate lyase family protein2 from burkholderia cenocepacia j2315
67 c1odpA_
not modelled
19.1
44
PDB header: lipid transportChain: A: PDB Molecule: apoa-i peptide;PDBTitle: peptide of human apoa-i residues 166-185. nmr, 5 structures2 at ph 6.6, 37 degrees celsius and peptide:sds mole ratio3 of 1:40
68 c1odrA_
not modelled
19.1
44
PDB header: lipid transportChain: A: PDB Molecule: apoa-i peptide;PDBTitle: peptide of human apoa-i residues 166-185. nmr, 5 structures2 at ph 6.0, 37 degrees celsius and peptide:dpc mole ratio3 of 1:40
69 c1odqA_
not modelled
19.1
44
PDB header: lipid transportChain: A: PDB Molecule: apoa-i peptide;PDBTitle: peptide of human apoa-i residues 166-185. nmr, 5 structures2 at ph 3.7, 37 degrees celsius and peptide:sds mole ratio3 of 1:40
70 c5i92E_
not modelled
19.0
21
PDB header: isomeraseChain: E: PDB Molecule: glutamate-1-semialdehyde 2,1-aminomutase;PDBTitle: crystal structure of glutamate-1-semialdehyde 2,1- aminomutase (gsa)2 from pseudomonas aeruginosa
71 d2a13a1
not modelled
19.0
33
Fold: LipocalinsSuperfamily: LipocalinsFamily: Rv2717c-like
72 c3u88B_
not modelled
18.9
33
PDB header: transcriptionChain: B: PDB Molecule: menin;PDBTitle: crystal structure of human menin in complex with mll1 and ledgf
73 c4wwxB_
not modelled
18.7
22
PDB header: hydrolase, ligaseChain: B: PDB Molecule: v(d)j recombination-activating protein 1;PDBTitle: crystal structure of the core rag1/2 recombinase
74 d1gkza1
not modelled
18.5
23
Fold: Bromodomain-likeSuperfamily: alpha-ketoacid dehydrogenase kinase, N-terminal domainFamily: alpha-ketoacid dehydrogenase kinase, N-terminal domain
75 c6nd4a_
not modelled
18.5
23
PDB header: ribosomeChain: A: PDB Molecule: mpp10;PDBTitle: conformational switches control early maturation of the eukaryotic2 small ribosomal subunit
76 d2gjpa2
not modelled
18.4
29
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain
77 d1ubea2
not modelled
17.9
19
Fold: Anti-LPS factor/recA domainSuperfamily: RecA protein, C-terminal domainFamily: RecA protein, C-terminal domain
78 d1mo6a2
not modelled
17.8
14
Fold: Anti-LPS factor/recA domainSuperfamily: RecA protein, C-terminal domainFamily: RecA protein, C-terminal domain
79 c2obvA_
not modelled
17.4
17
PDB header: transferaseChain: A: PDB Molecule: s-adenosylmethionine synthetase isoform type-1;PDBTitle: crystal structure of the human s-adenosylmethionine synthetase 1 in2 complex with the product
80 c2uv1B_
not modelled
17.3
17
PDB header: inhibitorChain: B: PDB Molecule: host-nuclease inhibitor protein gam;PDBTitle: hexagonal crystal form of gams from bacteriophage lambda.
81 c3viqC_
not modelled
17.2
20
PDB header: recombination activatorChain: C: PDB Molecule: swi5-dependent recombination dna repair protein 1;PDBTitle: crystal structure of swi5-sfr1 complex from fission yeast
82 c3fzvC_
not modelled
17.2
10
PDB header: transcription regulatorChain: C: PDB Molecule: probable transcriptional regulator;PDBTitle: crystal structure of pa01 protein, putative lysr family2 transcriptional regulator from pseudomonas aeruginosa
83 c3ia8A_
not modelled
16.8
8
PDB header: metal binding proteinChain: A: PDB Molecule: thap domain-containing protein 4;PDBTitle: the structure of the c-terminal heme nitrobindin domain of thap2 domain-containing protein 4 from homo sapiens
84 c3ispA_
not modelled
16.7
11
PDB header: transcriptionChain: A: PDB Molecule: hth-type transcriptional regulatorPDBTitle: crystal structure of argp from mycobacterium tuberculosis
85 c5vtmX_
not modelled
16.7
13
PDB header: transport proteinChain: X: PDB Molecule: type ii secretion system protein k;PDBTitle: the crystal structure of minor pseudopilin ternary complex of xcpvwx2 from the type 2 secretion system of pseudomonas aeruginosa
86 c4akkA_
not modelled
16.4
22
PDB header: transcriptionChain: A: PDB Molecule: nitrate regulatory protein;PDBTitle: structure of the nasr transcription antiterminator
87 c3qd7X_
not modelled
16.3
9
PDB header: hydrolaseChain: X: PDB Molecule: uncharacterized protein ydal;PDBTitle: crystal structure of ydal, a stand-alone small muts-related protein2 from escherichia coli
88 c2ideE_
not modelled
16.1
18
PDB header: biosynthetic proteinChain: E: PDB Molecule: molybdenum cofactor biosynthesis protein c;PDBTitle: crystal structure of the molybdenum cofactor biosynthesis protein c2 (ttha1789) from thermus theromophilus hb8
89 c6dk9I_
not modelled
16.1
15
PDB header: lyaseChain: I: PDB Molecule: dna damage-inducible protein;PDBTitle: yeast ddi2 cyanamide hydratase
90 c5tf0B_
not modelled
16.1
15
PDB header: hydrolaseChain: B: PDB Molecule: glycosyl hydrolase family 3 n-terminal domain protein;PDBTitle: crystal structure of glycosil hydrolase family 3 n-terminal domain2 protein from bacteroides intestinalis
91 c5z4yB_
not modelled
16.0
10
PDB header: dna binding proteinChain: B: PDB Molecule: cys regulon transcriptional activator;PDBTitle: crystal structure of pacysb ntd domain with space group p4
92 c5xfsB_
not modelled
16.0
15
PDB header: protein transportChain: B: PDB Molecule: ppe family protein ppe15;PDBTitle: crystal structure of pe8-ppe15 in complex with espg5 from m.2 tuberculosis
93 c5y9sD_
not modelled
15.9
7
PDB header: transcriptionChain: D: PDB Molecule: vv2_1132;PDBTitle: crystal structure of vv2_1132, a lysr family transcriptional regulator
94 c3cq0B_
not modelled
15.7
13
PDB header: transferaseChain: B: PDB Molecule: putative transaldolase ygr043c;PDBTitle: crystal structure of tal2_yeast
95 c5ed9A_
not modelled
15.7
26
PDB header: transport proteinChain: A: PDB Molecule: sun domain-containing protein 2;PDBTitle: crystal structure of cc1 of mouse sun2
96 c3so4C_
not modelled
15.4
16
PDB header: transferaseChain: C: PDB Molecule: methionine-adenosyltransferase;PDBTitle: methionine-adenosyltransferase from entamoeba histolytica
97 c5ydcA_
not modelled
15.3
13
PDB header: transcription regulatorChain: A: PDB Molecule: uncharacterized hth-type transcriptional regulator rv1828;PDBTitle: crystal structure of mercury soaked c-terminal domain of rv1828 from2 mycobacterium tuberculosis
98 c4e0uB_
not modelled
15.1
23
PDB header: transferaseChain: B: PDB Molecule: cyclic dipeptide n-prenyltransferase;PDBTitle: crystal structure of cdpnpt in complex with thiolodiphosphate and (s)-2 benzodiazependione
99 c3ghpA_
not modelled
15.0
50
PDB header: structural proteinChain: A: PDB Molecule: cellulosomal scaffoldin adaptor protein b;PDBTitle: structure of the second type ii cohesin module from the adaptor scaa2 scaffoldin of acetivibrio cellulolyticus (including long c-terminal3 linker)