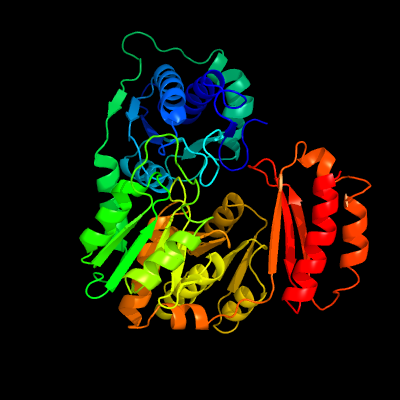
| 1 | c3pdkB_
|
|
 |
100.0 |
48 |
PDB header:isomerase
Chain: B: PDB Molecule:phosphoglucosamine mutase;
PDBTitle: crystal structure of phosphoglucosamine mutase from b. anthracis
|
|
|
|
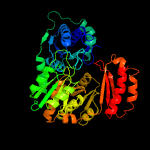
| 2 | c6gyzB_
|
|
 |
100.0 |
47 |
PDB header:isomerase
Chain: B: PDB Molecule:phosphoglucosamine mutase;
PDBTitle: crystal structure of glmm from staphylococcus aureus
|
|
|
|
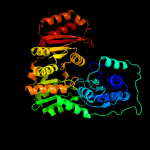
| 3 | c1wqaB_
|
|
 |
100.0 |
35 |
PDB header:isomerase
Chain: B: PDB Molecule:phospho-sugar mutase;
PDBTitle: crystal structure of pyrococcus horikoshii2 phosphomannomutase/phosphoglucomutase complexed with mg2+
|
|
|
|
| 4 | c2f7lA_
|
|
 |
100.0 |
29 |
PDB header:isomerase
Chain: A: PDB Molecule:455aa long hypothetical phospho-sugar mutase;
PDBTitle: crystal structure of sulfolobus tokodaii2 phosphomannomutase/phosphoglucomutase
|
|
|
|
| 5 | c3i3wB_
|
|
 |
100.0 |
37 |
PDB header:isomerase
Chain: B: PDB Molecule:phosphoglucosamine mutase;
PDBTitle: structure of a phosphoglucosamine mutase from francisella tularensis
|
|
|
|
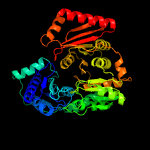
| 6 | c3c04A_
|
|
 |
100.0 |
27 |
PDB header:isomerase
Chain: A: PDB Molecule:phosphomannomutase/phosphoglucomutase;
PDBTitle: structure of the p368g mutant of pmm/pgm from p. aeruginosa
|
|
|
|
| 7 | c3uw2A_
|
|
 |
100.0 |
26 |
PDB header:isomerase
Chain: A: PDB Molecule:phosphoglucomutase/phosphomannomutase family protein;
PDBTitle: x-ray crystal structure of phosphoglucomutase/phosphomannomutase2 family protein (bth_i1489)from burkholderia thailandensis
|
|
|
|
| 8 | c5bmpA_
|
|
 |
100.0 |
25 |
PDB header:isomerase
Chain: A: PDB Molecule:phosphoglucomutase;
PDBTitle: crystal structure of phosphoglucomutase from xanthomonas citri2 complexed with glucose-1-phosphate
|
|
|
|
| 9 | c2fuvB_
|
|
 |
100.0 |
23 |
PDB header:isomerase
Chain: B: PDB Molecule:phosphoglucomutase;
PDBTitle: phosphoglucomutase from salmonella typhimurium.
|
|
|
|
| 10 | c1tuoA_
|
|
 |
100.0 |
25 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:putative phosphomannomutase;
PDBTitle: crystal structure of putative phosphomannomutase from2 thermus thermophilus hb8
|
|
|
|
| 11 | c1c4gB_
|
|
 |
100.0 |
20 |
PDB header:transferase
Chain: B: PDB Molecule:protein (alpha-d-glucose 1-phosphate phosphoglucomutase);
PDBTitle: phosphoglucomutase vanadate based transition state analog complex
|
|
|
|
| 12 | c4hjhA_
|
|
 |
100.0 |
26 |
PDB header:isomerase
Chain: A: PDB Molecule:phosphomannomutase;
PDBTitle: iodide sad phased crystal structure of a phosphoglucomutase from2 brucella melitensis complexed with glucose-6-phosphate
|
|
|
|
| 13 | c2z0fA_
|
|
 |
100.0 |
24 |
PDB header:isomerase
Chain: A: PDB Molecule:putative phosphoglucomutase;
PDBTitle: crystal structure of putative phosphoglucomutase from thermus2 thermophilus hb8
|
|
|
|
| 14 | c1kfiA_
|
|
 |
100.0 |
21 |
PDB header:isomerase
Chain: A: PDB Molecule:phosphoglucomutase 1;
PDBTitle: crystal structure of the exocytosis-sensitive2 phosphoprotein, pp63/parafusin (phosphoglucomutase) from3 paramecium
|
|
|
|
| 15 | c4qg5D_
|
|
 |
100.0 |
21 |
PDB header:isomerase
Chain: D: PDB Molecule:putative phosphoglucomutase;
PDBTitle: crystal structure of phosphoglucomutase from leishmania major at 3.52 angstrom resolution
|
|
|
|
| 16 | c2dkdA_
|
|
 |
100.0 |
23 |
PDB header:isomerase
Chain: A: PDB Molecule:phosphoacetylglucosamine mutase;
PDBTitle: crystal structure of n-acetylglucosamine-phosphate mutase,2 a member of the alpha-d-phosphohexomutase superfamily, in3 the product complex
|
|
|
|
| 17 | c4bjuB_
|
|
 |
100.0 |
25 |
PDB header:isomerase
Chain: B: PDB Molecule:n-acetylglucosamine-phosphate mutase;
PDBTitle: genetic and structural validation of aspergillus fumigatus2 n-acetylphosphoglucosamine mutase as an antifungal target
|
|
|
|
| 18 | d1p5dx1
|
|
 |
100.0 |
31 |
Fold:Phosphoglucomutase, first 3 domains
Superfamily:Phosphoglucomutase, first 3 domains
Family:Phosphoglucomutase, first 3 domains |
|
|
|
| 19 | d1kfia1
|
|
 |
100.0 |
22 |
Fold:Phosphoglucomutase, first 3 domains
Superfamily:Phosphoglucomutase, first 3 domains
Family:Phosphoglucomutase, first 3 domains |
|
|
|
| 20 | d3pmga1
|
|
 |
100.0 |
20 |
Fold:Phosphoglucomutase, first 3 domains
Superfamily:Phosphoglucomutase, first 3 domains
Family:Phosphoglucomutase, first 3 domains |
|
|
|
| 21 | d1p5dx3 |
|
not modelled |
99.9 |
19 |
Fold:Phosphoglucomutase, first 3 domains
Superfamily:Phosphoglucomutase, first 3 domains
Family:Phosphoglucomutase, first 3 domains |
|
|
| 22 | d1kfia3 |
|
not modelled |
99.9 |
20 |
Fold:Phosphoglucomutase, first 3 domains
Superfamily:Phosphoglucomutase, first 3 domains
Family:Phosphoglucomutase, first 3 domains |
|
|
| 23 | d1p5dx2 |
|
not modelled |
99.9 |
36 |
Fold:Phosphoglucomutase, first 3 domains
Superfamily:Phosphoglucomutase, first 3 domains
Family:Phosphoglucomutase, first 3 domains |
|
|
| 24 | d3pmga3 |
|
not modelled |
99.9 |
14 |
Fold:Phosphoglucomutase, first 3 domains
Superfamily:Phosphoglucomutase, first 3 domains
Family:Phosphoglucomutase, first 3 domains |
|
|
| 25 | d3pmga2 |
|
not modelled |
99.8 |
26 |
Fold:Phosphoglucomutase, first 3 domains
Superfamily:Phosphoglucomutase, first 3 domains
Family:Phosphoglucomutase, first 3 domains |
|
|
| 26 | d1kfia2 |
|
not modelled |
99.8 |
25 |
Fold:Phosphoglucomutase, first 3 domains
Superfamily:Phosphoglucomutase, first 3 domains
Family:Phosphoglucomutase, first 3 domains |
|
|
| 27 | d1p5dx4 |
|
not modelled |
99.7 |
19 |
Fold:TBP-like
Superfamily:Phosphoglucomutase, C-terminal domain
Family:Phosphoglucomutase, C-terminal domain |
|
|
| 28 | d1wjwa_ |
|
not modelled |
99.5 |
33 |
Fold:TBP-like
Superfamily:Phosphoglucomutase, C-terminal domain
Family:Phosphoglucomutase, C-terminal domain |
|
|
| 29 | d3pmga4 |
|
not modelled |
97.5 |
27 |
Fold:TBP-like
Superfamily:Phosphoglucomutase, C-terminal domain
Family:Phosphoglucomutase, C-terminal domain |
|
|
| 30 | d1kfia4 |
|
not modelled |
97.5 |
24 |
Fold:TBP-like
Superfamily:Phosphoglucomutase, C-terminal domain
Family:Phosphoglucomutase, C-terminal domain |
|
|
| 31 | d3bula2 |
|
not modelled |
91.4 |
15 |
Fold:Flavodoxin-like
Superfamily:Cobalamin (vitamin B12)-binding domain
Family:Cobalamin (vitamin B12)-binding domain |
|
|
| 32 | d1vmea1 |
|
not modelled |
91.2 |
16 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:Flavodoxin-related |
|
|
| 33 | d1nn4a_ |
|
not modelled |
91.2 |
26 |
Fold:Ribose/Galactose isomerase RpiB/AlsB
Superfamily:Ribose/Galactose isomerase RpiB/AlsB
Family:Ribose/Galactose isomerase RpiB/AlsB |
|
|
| 34 | c3s5pA_ |
|
not modelled |
91.1 |
15 |
PDB header:isomerase
Chain: A: PDB Molecule:ribose 5-phosphate isomerase;
PDBTitle: crystal structure of ribose-5-phosphate isomerase b rpib from giardia2 lamblia
|
|
|
| 35 | c3he8A_ |
|
not modelled |
91.0 |
21 |
PDB header:isomerase
Chain: A: PDB Molecule:ribose-5-phosphate isomerase;
PDBTitle: structural study of clostridium thermocellum ribose-5-phosphate2 isomerase b
|
|
|
| 36 | c3k7pA_ |
|
not modelled |
88.5 |
24 |
PDB header:isomerase
Chain: A: PDB Molecule:ribose 5-phosphate isomerase;
PDBTitle: structure of mutant of ribose 5-phosphate isomerase type b from2 trypanosoma cruzi.
|
|
|
| 37 | c3m1pA_ |
|
not modelled |
88.5 |
24 |
PDB header:isomerase
Chain: A: PDB Molecule:ribose 5-phosphate isomerase;
PDBTitle: structure of ribose 5-phosphate isomerase type b from trypanosoma2 cruzi, soaked with allose-6-phosphate
|
|
|
| 38 | c1k98A_ |
|
not modelled |
87.7 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:methionine synthase;
PDBTitle: adomet complex of meth c-terminal fragment
|
|
|
| 39 | c4m6iA_ |
|
not modelled |
86.1 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:peptidoglycan amidase rv3717;
PDBTitle: structure of the reduced, zn-bound form of mycobacterium tuberculosis2 peptidoglycan amidase rv3717
|
|
|
| 40 | c3qayC_ |
|
not modelled |
85.0 |
18 |
PDB header:lyase
Chain: C: PDB Molecule:endolysin;
PDBTitle: catalytic domain of cd27l endolysin targeting clostridia difficile
|
|
|
| 41 | c3hlyA_ |
|
not modelled |
82.4 |
19 |
PDB header:flavoprotein
Chain: A: PDB Molecule:flavodoxin-like domain;
PDBTitle: crystal structure of the flavodoxin-like domain from synechococcus sp2 q5mzp6_synp6 protein. northeast structural genomics consortium target3 snr135d.
|
|
|
| 42 | c4lfnD_ |
|
not modelled |
81.6 |
18 |
PDB header:isomerase
Chain: D: PDB Molecule:galactose-6-phosphate isomerase subunit b;
PDBTitle: crystal structure of d-galactose-6-phosphate isomerase in complex with2 d-ribulose
|
|
|
| 43 | c4em8A_ |
|
not modelled |
81.4 |
24 |
PDB header:isomerase
Chain: A: PDB Molecule:ribose 5-phosphate isomerase b;
PDBTitle: the structure of ribose 5-phosphate isomerase b from anaplasma2 phagocytophilum
|
|
|
| 44 | c3whpA_ |
|
not modelled |
81.1 |
25 |
PDB header:gene regulation
Chain: A: PDB Molecule:probable transcriptional regulator;
PDBTitle: crystal structure of the c-terminal domain of themus thermophilus litr2 in complex with cobalamin
|
|
|
| 45 | d1ccwa_ |
|
not modelled |
80.8 |
18 |
Fold:Flavodoxin-like
Superfamily:Cobalamin (vitamin B12)-binding domain
Family:Cobalamin (vitamin B12)-binding domain |
|
|
| 46 | c1y80A_ |
|
not modelled |
80.2 |
24 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:predicted cobalamin binding protein;
PDBTitle: structure of a corrinoid (factor iiim)-binding protein from moorella2 thermoacetica
|
|
|
| 47 | d1ycga1 |
|
not modelled |
79.2 |
22 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:Flavodoxin-related |
|
|
| 48 | d2vvpa1 |
|
not modelled |
78.7 |
30 |
Fold:Ribose/Galactose isomerase RpiB/AlsB
Superfamily:Ribose/Galactose isomerase RpiB/AlsB
Family:Ribose/Galactose isomerase RpiB/AlsB |
|
|
| 49 | c4lfmA_ |
|
not modelled |
78.4 |
21 |
PDB header:isomerase
Chain: A: PDB Molecule:galactose-6-phosphate isomerase subunit a;
PDBTitle: crystal structure of d-galactose-6-phosphate isomerase in complex with2 d-psicose
|
|
|
| 50 | c3fniA_ |
|
not modelled |
78.3 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative diflavin flavoprotein a 3;
PDBTitle: crystal structure of a diflavin flavoprotein a3 (all3895) from nostoc2 sp., northeast structural genomics consortium target nsr431a
|
|
|
| 51 | c2i2xD_ |
|
not modelled |
78.1 |
13 |
PDB header:transferase
Chain: D: PDB Molecule:methyltransferase 1;
PDBTitle: crystal structure of methanol:cobalamin methyltransferase complex2 mtabc from methanosarcina barkeri
|
|
|
| 52 | c6mu0A_ |
|
not modelled |
76.8 |
14 |
PDB header:isomerase
Chain: A: PDB Molecule:probable ribose-5-phosphate isomerase b;
PDBTitle: crystal structure of ribose-5-phosphate isomerase b from mycoplasma2 genitalium with bound ribulose-5-phosphate
|
|
|
| 53 | c1bmtB_ |
|
not modelled |
75.5 |
14 |
PDB header:methyltransferase
Chain: B: PDB Molecule:methionine synthase;
PDBTitle: how a protein binds b12: a 3.o angstrom x-ray structure of2 the b12-binding domains of methionine synthase
|
|
|
| 54 | c3ne8A_ |
|
not modelled |
74.1 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:n-acetylmuramoyl-l-alanine amidase;
PDBTitle: the crystal structure of a domain from n-acetylmuramoyl-l-alanine2 amidase of bartonella henselae str. houston-1
|
|
|
| 55 | d1xrsb1 |
|
not modelled |
72.5 |
16 |
Fold:Flavodoxin-like
Superfamily:Cobalamin (vitamin B12)-binding domain
Family:Cobalamin (vitamin B12)-binding domain |
|
|
| 56 | c3ezxA_ |
|
not modelled |
70.8 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:monomethylamine corrinoid protein 1;
PDBTitle: structure of methanosarcina barkeri monomethylamine2 corrinoid protein
|
|
|
| 57 | c5emiA_ |
|
not modelled |
67.9 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:cell wall hydrolase/autolysin;
PDBTitle: n-acetylmuramoyl-l-alanine amidase amic2 of nostoc punctiforme
|
|
|
| 58 | c4jgiB_ |
|
not modelled |
63.3 |
15 |
PDB header:protein binding
Chain: B: PDB Molecule:putative uncharacterized protein;
PDBTitle: 1.5 angstrom crystal structure of a novel cobalamin-binding protein2 from desulfitobacterium hafniense dcb-2
|
|
|
| 59 | d1o1xa_ |
|
not modelled |
63.2 |
23 |
Fold:Ribose/Galactose isomerase RpiB/AlsB
Superfamily:Ribose/Galactose isomerase RpiB/AlsB
Family:Ribose/Galactose isomerase RpiB/AlsB |
|
|
| 60 | c6apeA_ |
|
not modelled |
56.8 |
17 |
PDB header:oxidoreductase, hydrolase
Chain: A: PDB Molecule:bifunctional protein fold;
PDBTitle: crystal structure of bifunctional protein fold from helicobacter2 pylori
|
|
|
| 61 | c1xrsB_ |
|
not modelled |
56.2 |
17 |
PDB header:isomerase
Chain: B: PDB Molecule:d-lysine 5,6-aminomutase beta subunit;
PDBTitle: crystal structure of lysine 5,6-aminomutase in complex with plp,2 cobalamin, and 5'-deoxyadenosine
|
|
|
| 62 | c2ppwA_ |
|
not modelled |
56.1 |
15 |
PDB header:isomerase
Chain: A: PDB Molecule:conserved domain protein;
PDBTitle: the crystal structure of uncharacterized ribose 5-phosphate isomerase2 rpib from streptococcus pneumoniae
|
|
|
| 63 | c3qd5B_ |
|
not modelled |
54.6 |
28 |
PDB header:isomerase
Chain: B: PDB Molecule:putative ribose-5-phosphate isomerase;
PDBTitle: crystal structure of a putative ribose-5-phosphate isomerase from2 coccidioides immitis solved by combined iodide ion sad and mr
|
|
|
| 64 | c2yh5A_ |
|
not modelled |
53.9 |
11 |
PDB header:lipid binding protein
Chain: A: PDB Molecule:dapx protein;
PDBTitle: structure of the c-terminal domain of bamc
|
|
|
| 65 | d7reqa2 |
|
not modelled |
53.1 |
11 |
Fold:Flavodoxin-like
Superfamily:Cobalamin (vitamin B12)-binding domain
Family:Cobalamin (vitamin B12)-binding domain |
|
|
| 66 | d2a5la1 |
|
not modelled |
52.6 |
14 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:WrbA-like |
|
|
| 67 | d1d9ea_ |
|
not modelled |
48.2 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I DAHP synthetase |
|
|
| 68 | c5c8eC_ |
|
not modelled |
46.5 |
25 |
PDB header:transcription regulator/dna
Chain: C: PDB Molecule:light-dependent transcriptional regulator carh;
PDBTitle: crystal structure of thermus thermophilus carh bound to2 adenosylcobalamin and a 26-bp dna segment
|
|
|
| 69 | c3nvtA_ |
|
not modelled |
45.9 |
19 |
PDB header:transferase/isomerase
Chain: A: PDB Molecule:3-deoxy-d-arabino-heptulosonate 7-phosphate synthase;
PDBTitle: 1.95 angstrom crystal structure of a bifunctional 3-deoxy-7-2 phosphoheptulonate synthase/chorismate mutase (aroa) from listeria3 monocytogenes egd-e
|
|
|
| 70 | d1e5da1 |
|
not modelled |
44.0 |
17 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:Flavodoxin-related |
|
|
| 71 | c2yxbA_ |
|
not modelled |
42.0 |
17 |
PDB header:isomerase
Chain: A: PDB Molecule:coenzyme b12-dependent mutase;
PDBTitle: crystal structure of the methylmalonyl-coa mutase alpha-subunit from2 aeropyrum pernix
|
|
|
| 72 | d1ml4a2 |
|
not modelled |
41.2 |
13 |
Fold:ATC-like
Superfamily:Aspartate/ornithine carbamoyltransferase
Family:Aspartate/ornithine carbamoyltransferase |
|
|
| 73 | d1b1ca_ |
|
not modelled |
40.7 |
9 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:Cytochrome p450 reductase N-terminal domain-like |
|
|
| 74 | c1vmeB_ |
|
not modelled |
40.6 |
12 |
PDB header:electron transport
Chain: B: PDB Molecule:flavoprotein;
PDBTitle: crystal structure of flavoprotein (tm0755) from thermotoga maritima at2 1.80 a resolution
|
|
|
| 75 | c3f93D_ |
|
not modelled |
40.1 |
22 |
PDB header:hydrolase
Chain: D: PDB Molecule:beta-glucosidase;
PDBTitle: crystal structure of exo-1,3/1,4-beta-glucanase (exop) from2 pseudoalteromonas sp. bb1
|
|
|
| 76 | c6fxwA_ |
|
not modelled |
39.2 |
22 |
PDB header:isomerase
Chain: A: PDB Molecule:putative ribose 5-phosphate isomerase;
PDBTitle: structure of leishmania infantum type b ribose 5-phosphate isomerase
|
|
|
| 77 | c3qqzA_ |
|
not modelled |
37.7 |
20 |
PDB header:metal binding protein
Chain: A: PDB Molecule:putative uncharacterized protein yjik;
PDBTitle: crystal structure of the c-terminal domain of the yjik protein from2 escherichia coli cft073
|
|
|
| 78 | c3sz8D_ |
|
not modelled |
36.0 |
19 |
PDB header:transferase
Chain: D: PDB Molecule:2-dehydro-3-deoxyphosphooctonate aldolase 2;
PDBTitle: crystal structure of 2-dehydro-3-deoxyphosphooctonate aldolase from2 burkholderia pseudomallei
|
|
|
| 79 | c4binA_ |
|
not modelled |
35.0 |
26 |
PDB header:hydrolase
Chain: A: PDB Molecule:n-acetylmuramoyl-l-alanine amidase amic;
PDBTitle: crystal structure of the e. coli n-acetylmuramoyl-l-alanine amidase2 amic
|
|
|
| 80 | c5nbsA_ |
|
not modelled |
34.9 |
20 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-glucosidase;
PDBTitle: structural studies of a glycoside hydrolase family 3 beta-glucosidase2 from the model fungus neurospora crassa
|
|
|
| 81 | c2ohiB_ |
|
not modelled |
34.6 |
14 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:type a flavoprotein fpra;
PDBTitle: crystal structure of coenzyme f420h2 oxidase (fpra), a diiron2 flavoprotein, reduced state
|
|
|
| 82 | d1xk7a1 |
|
not modelled |
34.0 |
38 |
Fold:CoA-transferase family III (CaiB/BaiF)
Superfamily:CoA-transferase family III (CaiB/BaiF)
Family:CoA-transferase family III (CaiB/BaiF) |
|
|
| 83 | c6mdyC_ |
|
not modelled |
33.5 |
18 |
PDB header:hydrolase
Chain: C: PDB Molecule:2-dehydro-3-deoxyphosphooctonate aldolase;
PDBTitle: crystal structure of a 2-dehydro-3-deoxyphosphooctonate aldolase from2 legionella pneumophila philadelphia 1
|
|
|
| 84 | c4xfrB_ |
|
not modelled |
33.2 |
20 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of a domain of unknown function (duf1537) from2 bordetella bronchiseptica (bb3215), target efi-511620, with bound3 citrate, domain swapped dimer, space group p6522
|
|
|
| 85 | c2q9uB_ |
|
not modelled |
32.6 |
13 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:a-type flavoprotein;
PDBTitle: crystal structure of the flavodiiron protein from giardia2 intestinalis
|
|
|
| 86 | c4amuB_ |
|
not modelled |
31.9 |
13 |
PDB header:transferase
Chain: B: PDB Molecule:ornithine carbamoyltransferase, catabolic;
PDBTitle: structure of ornithine carbamoyltransferase from mycoplasma2 penetrans with a p321 space group
|
|
|
| 87 | c5mp4C_ |
|
not modelled |
31.9 |
13 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:protoplast secreted protein 2;
PDBTitle: the structure of pst2p from saccharomyces cerevisiae
|
|
|
| 88 | c3tqkA_ |
|
not modelled |
31.5 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:phospho-2-dehydro-3-deoxyheptonate aldolase;
PDBTitle: structure of phospho-2-dehydro-3-deoxyheptonate aldolase from2 francisella tularensis schu s4
|
|
|
| 89 | c2pfsA_ |
|
not modelled |
31.3 |
18 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:universal stress protein;
PDBTitle: crystal structure of universal stress protein from nitrosomonas2 europaea
|
|
|
| 90 | d1dxha2 |
|
not modelled |
30.9 |
14 |
Fold:ATC-like
Superfamily:Aspartate/ornithine carbamoyltransferase
Family:Aspartate/ornithine carbamoyltransferase |
|
|
| 91 | d1ak2a1 |
|
not modelled |
30.5 |
10 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nucleotide and nucleoside kinases |
|
|
| 92 | c2zkiH_ |
|
not modelled |
30.4 |
15 |
PDB header:transcription
Chain: H: PDB Molecule:199aa long hypothetical trp repressor binding
PDBTitle: crystal structure of hypothetical trp repressor binding2 protein from sul folobus tokodaii (st0872)
|
|
|
| 93 | c1xa3B_ |
|
not modelled |
30.1 |
38 |
PDB header:transferase
Chain: B: PDB Molecule:crotonobetainyl-coa:carnitine coa-transferase;
PDBTitle: crystal structure of caib, a type iii coa transferase in2 carnitine metabolism
|
|
|
| 94 | c4heqB_ |
|
not modelled |
30.1 |
25 |
PDB header:electron transport
Chain: B: PDB Molecule:flavodoxin;
PDBTitle: the crystal structure of flavodoxin from desulfovibrio gigas
|
|
|
| 95 | c4khaA_ |
|
not modelled |
28.7 |
13 |
PDB header:chaperone/nuclear protein
Chain: A: PDB Molecule:spt16m-histone h2b 1.1 chimera;
PDBTitle: structural basis of histone h2a-h2b recognition by the essential2 chaperone fact
|
|
|
| 96 | c4r3uD_ |
|
not modelled |
28.4 |
17 |
PDB header:isomerase
Chain: D: PDB Molecule:2-hydroxyisobutyryl-coa mutase small subunit;
PDBTitle: crystal structure of 2-hydroxyisobutyryl-coa mutase
|
|
|
| 97 | c1a4iB_ |
|
not modelled |
28.3 |
13 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:methylenetetrahydrofolate dehydrogenase /
PDBTitle: human tetrahydrofolate dehydrogenase / cyclohydrolase
|
|
|
| 98 | d1kjna_ |
|
not modelled |
27.9 |
24 |
Fold:Hypothetical protein MTH777 (MT0777)
Superfamily:Hypothetical protein MTH777 (MT0777)
Family:Hypothetical protein MTH777 (MT0777) |
|
|
| 99 | c3eglC_ |
|
not modelled |
27.7 |
16 |
PDB header:structural genomics, unknown function
Chain: C: PDB Molecule:degv family protein;
PDBTitle: crystal structure of degv family protein cg2579 from corynebacterium2 glutamicum
|
|
|
| 100 | c4xz6A_ |
|
not modelled |
27.1 |
12 |
PDB header:transport protein
Chain: A: PDB Molecule:glycine betaine/proline abc transporter, periplasmic
PDBTitle: tmox in complex with tmao
|
|
|
| 101 | c3nolA_ |
|
not modelled |
27.0 |
24 |
PDB header:transferase
Chain: A: PDB Molecule:glutamine cyclotransferase;
PDBTitle: crystal structure of zymomonas mobilis glutaminyl cyclase (trigonal2 form)
|
|
|
| 102 | d1f4pa_ |
|
not modelled |
26.8 |
11 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:Flavodoxin-related |
|
|
| 103 | c5o5oD_ |
|
not modelled |
26.1 |
27 |
PDB header:chaperone
Chain: D: PDB Molecule:rnase adapter protein rapz;
PDBTitle: x-ray crystal structure of rapz from escherichia coli (p32 space2 group)
|
|
|
| 104 | c5vegC_ |
|
not modelled |
25.8 |
18 |
PDB header:electron transport
Chain: C: PDB Molecule:flavodoxin;
PDBTitle: structure of a short-chain flavodoxin associated with a non-canonical2 pdu bacterial microcompartment
|
|
|
| 105 | c6h0cA_ |
|
not modelled |
25.7 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative diflavin flavoprotein a 3;
PDBTitle: flv1 flavodiiron core from synechocystis sp. pcc6803
|
|
|
| 106 | d1q7ea_ |
|
not modelled |
25.7 |
24 |
Fold:CoA-transferase family III (CaiB/BaiF)
Superfamily:CoA-transferase family III (CaiB/BaiF)
Family:CoA-transferase family III (CaiB/BaiF) |
|
|
| 107 | c4h2dB_ |
|
not modelled |
25.5 |
7 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:nadph-dependent diflavin oxidoreductase 1;
PDBTitle: crystal structure of ndor1
|
|
|
| 108 | c2g04B_ |
|
not modelled |
25.1 |
19 |
PDB header:isomerase
Chain: B: PDB Molecule:probable fatty-acid-coa racemase far;
PDBTitle: crystal structure of fatty acid-coa racemase from mycobacterium2 tuberculosis h37rv
|
|
|
| 109 | d1q77a_ |
|
not modelled |
25.1 |
13 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:Universal stress protein-like |
|
|
| 110 | c5z87B_ |
|
not modelled |
24.9 |
18 |
PDB header:hydrolase
Chain: B: PDB Molecule:emgh1;
PDBTitle: structural of a novel b-glucosidase emgh1 at 2.3 angstrom from2 erythrobacter marinus
|
|
|
| 111 | d1ykga1 |
|
not modelled |
24.9 |
5 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:Cytochrome p450 reductase N-terminal domain-like |
|
|
| 112 | c5oa1V_ |
|
not modelled |
24.9 |
21 |
PDB header:transcription
Chain: V: PDB Molecule:rna polymerase i-specific transcription initiation factor
PDBTitle: rna polymerase i pre-initiation complex
|
|
|
| 113 | c1e5dA_ |
|
not modelled |
24.8 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:rubredoxin\:oxygen oxidoreductase;
PDBTitle: rubredoxin oxygen:oxidoreductase (roo) from anaerobe desulfovibrio2 gigas
|
|
|
| 114 | c4hh3C_ |
|
not modelled |
24.7 |
16 |
PDB header:flavoprotein/transcription
Chain: C: PDB Molecule:appa protein;
PDBTitle: structure of the appa-ppsr2 core complex from rb. sphaeroides
|
|
|
| 115 | d1fmfa_ |
|
not modelled |
24.3 |
20 |
Fold:Flavodoxin-like
Superfamily:Cobalamin (vitamin B12)-binding domain
Family:Cobalamin (vitamin B12)-binding domain |
|
|
| 116 | c4wnyA_ |
|
not modelled |
23.9 |
10 |
PDB header:signaling protein
Chain: A: PDB Molecule:universal stress protein;
PDBTitle: crystal structure of a protein from the universal stress protein2 family from burkholderia pseudomallei
|
|
|
| 117 | c2nz2A_ |
|
not modelled |
23.7 |
28 |
PDB header:ligase
Chain: A: PDB Molecule:argininosuccinate synthase;
PDBTitle: crystal structure of human argininosuccinate synthase in complex with2 aspartate and citrulline
|
|
|
| 118 | c4hl6D_ |
|
not modelled |
22.9 |
30 |
PDB header:transferase
Chain: D: PDB Molecule:uncharacterized protein yfde;
PDBTitle: yfde from escherichia coli
|
|
|
| 119 | c3s3tD_ |
|
not modelled |
22.7 |
5 |
PDB header:chaperone
Chain: D: PDB Molecule:nucleotide-binding protein, universal stress protein uspa
PDBTitle: universal stress protein uspa from lactobacillus plantarum
|
|
|
| 120 | d2ak3a1 |
|
not modelled |
22.6 |
9 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nucleotide and nucleoside kinases |
|
|