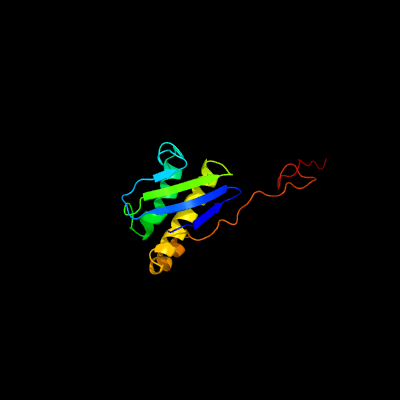
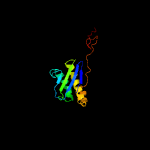
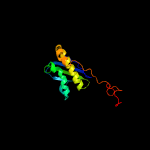
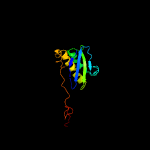
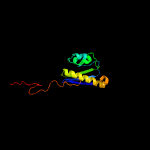
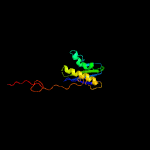
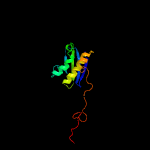
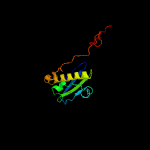
1 c5o5jI_
100.0
90
PDB header: ribosomeChain: I: PDB Molecule: 30s ribosomal protein s9;PDBTitle: structure of the 30s small ribosomal subunit from mycobacterium2 smegmatis
2 d2gy9i1
100.0
50
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: Translational machinery components
3 d2vqei1
100.0
52
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: Translational machinery components
4 c3bbnI_
100.0
53
PDB header: ribosomeChain: I: PDB Molecule: ribosomal protein s9;PDBTitle: homology model for the spinach chloroplast 30s subunit fitted to 9.4a2 cryo-em map of the 70s chlororibosome.
5 c3j6vI_
100.0
31
PDB header: ribosomeChain: I: PDB Molecule: 28s ribosomal protein s9, mitochondrial;PDBTitle: cryo-em structure of the small subunit of the mammalian mitochondrial2 ribosome
6 c2xzmI_
100.0
28
PDB header: ribosomeChain: I: PDB Molecule: rps16e;PDBTitle: crystal structure of the eukaryotic 40s ribosomal2 subunit in complex with initiation factor 1. this file3 contains the 40s subunit and initiation factor for4 molecule 1
7 c2zkqi_
100.0
33
PDB header: ribosomal protein/rnaChain: I: PDB Molecule: PDBTitle: structure of a mammalian ribosomal 40s subunit within an 80s complex2 obtained by docking homology models of the rna and proteins into an3 8.7 a cryo-em map
8 c3zeyK_
100.0
27
PDB header: ribosomeChain: K: PDB Molecule: 40s ribosomal protein s16, putative;PDBTitle: high-resolution cryo-electron microscopy structure of the trypanosoma2 brucei ribosome
9 c5xyiQ_
100.0
30
PDB header: ribosomeChain: Q: PDB Molecule: 40s ribosomal protein s16, putative;PDBTitle: small subunit of trichomonas vaginalis ribosome
10 c3j20K_
100.0
35
PDB header: ribosomeChain: K: PDB Molecule: 30s ribosomal protein s9p;PDBTitle: promiscuous behavior of proteins in archaeal ribosomes revealed by2 cryo-em: implications for evolution of eukaryotic ribosomes (30s3 ribosomal subunit)
11 c1s1hI_
100.0
34
PDB header: ribosomeChain: I: PDB Molecule: 40s ribosomal protein s16;PDBTitle: structure of the ribosomal 80s-eef2-sordarin complex from yeast2 obtained by docking atomic models for rna and protein components into3 a 11.7 a cryo-em map. this file, 1s1h, contains 40s subunit. the 60s4 ribosomal subunit is in file 1s1i.
12 c3iz6I_
100.0
32
PDB header: ribosomeChain: I: PDB Molecule: 40s ribosomal protein s16 (s9p);PDBTitle: localization of the small subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
13 c5ujaA_
79.1
22
PDB header: protein transportChain: A: PDB Molecule: bovine multidrug resistance protein 1 (mrp1),multidrugPDBTitle: cryo-em structure of bovine multidrug resistance protein 1 (mrp1)2 bound to leukotriene c4
14 c5uj9A_
78.7
22
PDB header: transport proteinChain: A: PDB Molecule: bovine multidrug resistance protein 1 (mrp1),multidrugPDBTitle: cryo-em structure of bovine multidrug resistance protein 1 (mrp1)
15 c6bhuA_
73.9
22
PDB header: transport proteinChain: A: PDB Molecule: multidrug resistance-associated protein 1;PDBTitle: cryo-em structure of atp-bound, outward-facing bovine multidrug2 resistance protein 1 (mrp1)
16 c3g5uB_
68.0
26
PDB header: membrane proteinChain: B: PDB Molecule: multidrug resistance protein 1a;PDBTitle: structure of p-glycoprotein reveals a molecular basis for2 poly-specific drug binding
17 c4ry2A_
67.8
29
PDB header: transport protein/hydrolaseChain: A: PDB Molecule: abc-type bacteriocin transporter;PDBTitle: crystal structure of the peptidase-containing abc transporter pcat1
18 c5mkkA_
67.1
23
PDB header: transport proteinChain: A: PDB Molecule: multidrug resistance abc transporter atp-binding andPDBTitle: crystal structure of the heterodimeric abc transporter tmrab, a2 homolog of the antigen translocation complex tap
19 c5mkkB_
66.5
18
PDB header: transport proteinChain: B: PDB Molecule: multidrug resistance abc transporter atp-binding andPDBTitle: crystal structure of the heterodimeric abc transporter tmrab, a2 homolog of the antigen translocation complex tap
20 c4f4cA_
65.0
28
PDB header: hydrolase,protein transportChain: A: PDB Molecule: multidrug resistance protein pgp-1;PDBTitle: the crystal structure of the multi-drug transporter
21 c3u5eh_
not modelled
62.4
20
PDB header: ribosomeChain: H: PDB Molecule: 60s ribosomal protein l9-a;PDBTitle: the structure of the eukaryotic ribosome at 3.0 a resolution. this2 entry contains proteins of the 60s subunit, ribosome a
22 c3j3bh_
not modelled
60.9
29
PDB header: ribosomeChain: H: PDB Molecule: 60s ribosomal protein l9;PDBTitle: structure of the human 60s ribosomal proteins
23 c6c0vA_
not modelled
59.7
28
PDB header: transport proteinChain: A: PDB Molecule: multidrug resistance protein 1;PDBTitle: molecular structure of human p-glycoprotein in the atp-bound, outward-2 facing conformation
24 c5tsiA_
not modelled
58.2
21
PDB header: membrane protein, hydrolaseChain: A: PDB Molecule: cystic fibrosis transmembrane conductance regulator;PDBTitle: structure of the cystic fibrosis transmembrane conductance regulator2 (cftr) from zebrafish
25 c1s3bB_
not modelled
57.2
41
PDB header: oxidoreductaseChain: B: PDB Molecule: amine oxidase [flavin-containing] b;PDBTitle: crystal structure of maob in complex with n-methyl-n-2 propargyl-1(r)-aminoindan
26 c3wmeA_
not modelled
53.7
26
PDB header: transport proteinChain: A: PDB Molecule: atp-binding cassette, sub-family b, member 1;PDBTitle: crystal structure of an inward-facing eukaryotic abc multidrug2 transporter
27 c4aa3A_
not modelled
52.6
30
PDB header: transport proteinChain: A: PDB Molecule: atp-binding cassette sub-family b member 10,PDBTitle: structure of the human mitochondrial abc transporter,2 abcb10 (plate form)
28 c5ochH_
not modelled
49.4
29
PDB header: hydrolaseChain: H: PDB Molecule: atp-binding cassette sub-family b member 8, mitochondrial;PDBTitle: the crystal structure of human abcb8 in an outward-facing state
29 d1d5ta1
not modelled
49.2
20
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: GDI-like N domain
30 c5ttkB_
not modelled
48.8
36
PDB header: oxidoreductaseChain: B: PDB Molecule: amine oxidase;PDBTitle: crystal structure of selenomethionine-incorporated nicotine2 oxidoreductase from pseudomonas putida
31 c2v6oA_
not modelled
48.7
14
PDB header: oxidoreductaseChain: A: PDB Molecule: thioredoxin glutathione reductase;PDBTitle: structure of schistosoma mansoni thioredoxin-glutathione2 reductase (smtgr)
32 c1v0jB_
not modelled
48.5
33
PDB header: isomeraseChain: B: PDB Molecule: udp-galactopyranose mutase;PDBTitle: udp-galactopyranose mutase from mycobacterium tuberculosis
33 d1wdda1
not modelled
47.1
28
Fold: TIM beta/alpha-barrelSuperfamily: RuBisCo, C-terminal domainFamily: RuBisCo, large subunit, C-terminal domain
34 c3j39h_
not modelled
46.3
29
PDB header: ribosomeChain: H: PDB Molecule: 60s ribosomal protein l9;PDBTitle: structure of the d. melanogaster 60s ribosomal proteins
35 c6c3oE_
not modelled
46.1
17
PDB header: transport proteinChain: E: PDB Molecule: atp-binding cassette sub-family c member 8;PDBTitle: cryo-em structure of human katp bound to atp and adp in quatrefoil2 form
36 c3zf7k_
not modelled
45.0
32
PDB header: ribosomeChain: K: PDB Molecule: 60s ribosomal protein l10, putative;PDBTitle: high-resolution cryo-electron microscopy structure of the trypanosoma2 brucei ribosome
37 c6j0zC_
not modelled
44.4
32
PDB header: oxidoreductaseChain: C: PDB Molecule: putative angucycline-like polyketide oxygenase;PDBTitle: crystal structure of alpk
38 c5l22B_
not modelled
43.4
26
PDB header: protein transportChain: B: PDB Molecule: abc transporter (hlyb subfamily);PDBTitle: prtd t1ss abc transporter
39 c2rrlA_
not modelled
42.7
20
PDB header: protein transportChain: A: PDB Molecule: flagellar hook-length control protein;PDBTitle: solution structure of the c-terminal domain of the flik
40 d1rbla1
not modelled
42.0
30
Fold: TIM beta/alpha-barrelSuperfamily: RuBisCo, C-terminal domainFamily: RuBisCo, large subunit, C-terminal domain
41 c1ltxR_
not modelled
40.9
20
PDB header: transferase/protein bindingChain: R: PDB Molecule: rab escort protein 1;PDBTitle: structure of rab escort protein-1 in complex with rab geranylgeranyl2 transferase and isoprenoid
42 c3cp8C_
not modelled
40.1
27
PDB header: oxidoreductaseChain: C: PDB Molecule: trna uridine 5-carboxymethylaminomethylPDBTitle: crystal structure of gida from chlorobium tepidum
43 c3cp2A_
not modelled
39.0
32
PDB header: oxidoreductaseChain: A: PDB Molecule: trna uridine 5-carboxymethylaminomethylPDBTitle: crystal structure of gida from e. coli
44 c2yl4A_
not modelled
39.0
29
PDB header: membrane proteinChain: A: PDB Molecule: atp-binding cassette sub-family b member 10,PDBTitle: structure of the human mitochondrial abc transporter, abcb10
45 c5u1dB_
not modelled
38.0
26
PDB header: transport proteinChain: B: PDB Molecule: antigen peptide transporter 2;PDBTitle: cryo-em structure of the human tap atp-binding cassette transporter
46 c4i58A_
not modelled
37.8
38
PDB header: oxidoreductaseChain: A: PDB Molecule: cyclohexylamine oxidase;PDBTitle: cyclohexylamine oxidase from brevibacterium oxydans ih-35a
47 c6on2A_
not modelled
37.6
19
PDB header: hydrolaseChain: A: PDB Molecule: atp-dependent protease la;PDBTitle: lon protease from yersinia pestis with y2853 substrate
48 c3qf4A_
not modelled
37.2
26
PDB header: transport proteinChain: A: PDB Molecule: abc transporter, atp-binding protein;PDBTitle: crystal structure of a heterodimeric abc transporter in its inward-2 facing conformation
49 c5ykfH_
not modelled
36.5
20
PDB header: membrane proteinChain: H: PDB Molecule: atp-binding cassette sub-family c member 8 isoform x2;PDBTitle: structure of pancreatic atp-sensitive potassium channel bound with2 glibenclamide and atpgammas (3d class1 at 4.33a)
50 c4a1eU_
not modelled
36.1
35
PDB header: ribosomeChain: U: PDB Molecule: rpl35;PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 5s rrna, 5.8s rrna3 and proteins of molecule 1
51 c3jskN_
not modelled
35.9
21
PDB header: biosynthetic proteinChain: N: PDB Molecule: cypbp37 protein;PDBTitle: thiazole synthase from neurospora crassa
52 c2hydB_
not modelled
35.6
21
PDB header: transport proteinChain: B: PDB Molecule: abc transporter homolog;PDBTitle: multidrug abc transporter sav1866
53 d2bcgg1
not modelled
35.2
17
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: GDI-like N domain
54 c3k7tB_
not modelled
34.1
40
PDB header: oxidoreductaseChain: B: PDB Molecule: 6-hydroxy-l-nicotine oxidase;PDBTitle: crystal structure of apo-form 6-hydroxy-l-nicotine oxidase, crystal2 form p3121
55 d1hyua1
not modelled
33.0
23
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: FAD/NAD-linked reductases, N-terminal and central domains
56 c5do7A_
not modelled
32.7
23
PDB header: transport proteinChain: A: PDB Molecule: atp-binding cassette sub-family g member 5;PDBTitle: crystal structure of the human sterol transporter abcg5/abcg8
57 c2uzzD_
not modelled
32.5
24
PDB header: oxidoreductaseChain: D: PDB Molecule: n-methyl-l-tryptophan oxidase;PDBTitle: x-ray structure of n-methyl-l-tryptophan oxidase (mtox)
58 c5wgyA_
not modelled
32.3
29
PDB header: oxidoreductase/oxidoreductase inhibitorChain: A: PDB Molecule: flavin-dependent halogenase;PDBTitle: crystal structure of mala' c112s/c128s, malbrancheamide b complex
59 c4cnjD_
not modelled
32.0
30
PDB header: oxidoreductaseChain: D: PDB Molecule: l-amino acid oxidase;PDBTitle: l-aminoacetone oxidase from streptococcus oligofermentans2 belongs to a new 3-domain family of bacterial flavoproteins
60 c5u71A_
not modelled
31.9
24
PDB header: membrane protein, hydrolaseChain: A: PDB Molecule: cystic fibrosis transmembrane conductance regulator;PDBTitle: structure of human cystic fibrosis transmembrane conductance regulator2 (cftr)
61 c2zxiC_
not modelled
31.7
38
PDB header: fad-binding proteinChain: C: PDB Molecule: trna uridine 5-carboxymethylaminomethyl modification enzymePDBTitle: structure of aquifex aeolicus gida in the form ii crystal
62 c6bznA_
not modelled
31.4
23
PDB header: flavoproteinChain: A: PDB Molecule: halogenase pltm;PDBTitle: crystal structure of halogenase pltm
63 c4y4nE_
not modelled
31.0
31
PDB header: biosynthetic proteinChain: E: PDB Molecule: putative ribose 1,5-bisphosphate isomerase;PDBTitle: thiazole synthase thi4 from methanococcus igneus
64 d1gesa1
not modelled
30.9
36
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: FAD/NAD-linked reductases, N-terminal and central domains
65 c3rhaA_
not modelled
29.1
32
PDB header: oxidoreductaseChain: A: PDB Molecule: putrescine oxidase;PDBTitle: the crystal structure of oxidoreductase from arthrobacter aurescens
66 c5wuaF_
not modelled
28.8
14
PDB header: transport proteinChain: F: PDB Molecule: sur1;PDBTitle: structure of a pancreatic atp-sensitive potassium channel
67 c4yshA_
not modelled
27.6
30
PDB header: oxidoreductaseChain: A: PDB Molecule: glycine oxidase;PDBTitle: crystal structure of glycine oxidase from geobacillus kaustophilus
68 c4ia6B_
not modelled
26.8
32
PDB header: immune systemChain: B: PDB Molecule: myosin-crossreactive antigen;PDBTitle: hydratase from lactobacillus acidophilus in a ligand bound form (la2 lah)
69 c2olnA_
not modelled
26.7
43
PDB header: oxidoreductaseChain: A: PDB Molecule: nikd protein;PDBTitle: nikd, an unusual amino acid oxidase essential for2 nikkomycin biosynthesis: closed form at 1.15 a resolution
70 c1ryiB_
not modelled
26.0
33
PDB header: oxidoreductaseChain: B: PDB Molecule: glycine oxidase;PDBTitle: structure of glycine oxidase with bound inhibitor glycolate
71 c3g05B_
not modelled
26.0
32
PDB header: rna binding proteinChain: B: PDB Molecule: trna uridine 5-carboxymethylaminomethyl modification enzymePDBTitle: crystal structure of n-terminal domain (2-550) of e.coli mnmg
72 c2qaeA_
not modelled
25.6
27
PDB header: oxidoreductaseChain: A: PDB Molecule: dihydrolipoyl dehydrogenase;PDBTitle: crystal structure analysis of trypanosoma cruzi lipoamide2 dehydrogenase
73 c6c87A_
not modelled
25.2
21
PDB header: protein transportChain: A: PDB Molecule: rab gdp dissociation inhibitor alpha;PDBTitle: crystal structure of rab gdp dissociation inhibitor alpha from2 naegleria fowleri
74 c1f8sA_
not modelled
25.1
15
PDB header: oxidoreductaseChain: A: PDB Molecule: l-amino acid oxidase;PDBTitle: crystal structure of l-amino acid oxidase from calloselasma2 rhodostoma, complexed with three molecules of o-aminobenzoate.
75 d1rrea_
not modelled
25.1
22
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: ATP-dependent protease Lon (La), catalytic domain
76 c2nvkX_
not modelled
24.4
32
PDB header: oxidoreductaseChain: X: PDB Molecule: thioredoxin reductase;PDBTitle: crystal structure of thioredoxin reductase from drosophila2 melanogaster
77 d1h6va1
not modelled
23.7
32
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: FAD/NAD-linked reductases, N-terminal and central domains
78 c5ez7A_
not modelled
23.1
23
PDB header: oxidoreductaseChain: A: PDB Molecule: flavoenzyme pa4991;PDBTitle: crystal structure of the fad dependent oxidoreductase pa4991 from2 pseudomonas aeruginosa
79 c1dxlC_
not modelled
23.0
32
PDB header: oxidoreductaseChain: C: PDB Molecule: dihydrolipoamide dehydrogenase;PDBTitle: dihydrolipoamide dehydrogenase of glycine decarboxylase2 from pisum sativum
80 c3ihgA_
not modelled
21.7
20
PDB header: flavoprotein, oxidoreductaseChain: A: PDB Molecule: rdme;PDBTitle: crystal structure of a ternary complex of aklavinone-11 hydroxylase2 with fad and aklavinone
81 c6bwtD_
not modelled
21.6
27
PDB header: oxidoreductaseChain: D: PDB Molecule: thioredoxin reductase;PDBTitle: 2.45 angstrom resolution crystal structure thioredoxin reductase from2 francisella tularensis.
82 c5ochF_
not modelled
21.6
32
PDB header: hydrolaseChain: F: PDB Molecule: atp-binding cassette sub-family b member 8, mitochondrial;PDBTitle: the crystal structure of human abcb8 in an outward-facing state
83 c3dmeB_
not modelled
21.5
38
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: conserved exported protein;PDBTitle: crystal structure of conserved exported protein from bordetella2 pertussis. northeast structural genomics target ber141
84 d2hzaa1
not modelled
21.0
22
Fold: Ribbon-helix-helixSuperfamily: Ribbon-helix-helixFamily: CopG-like
85 d2i0za1
not modelled
20.3
32
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: HI0933 N-terminal domain-like
86 c3m6aC_
not modelled
20.3
24
PDB header: hydrolaseChain: C: PDB Molecule: atp-dependent protease la 1;PDBTitle: crystal structure of bacillus subtilis lon c-terminal domain
87 d2gf3a1
not modelled
20.2
24
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: FAD-linked reductases, N-terminal domain
88 d1chua2
not modelled
20.0
26
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: Succinate dehydrogenase/fumarate reductase flavoprotein N-terminal domain
89 c5e7sG_
not modelled
19.9
19
PDB header: hydrolaseChain: G: PDB Molecule: lon protease;PDBTitle: hexameric structure of a lona protease domain in active state
90 d2gmha1
not modelled
19.9
33
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: FAD-linked reductases, N-terminal domain
91 d1seza1
not modelled
19.8
37
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: FAD-linked reductases, N-terminal domain
92 c2cfyB_
not modelled
19.8
35
PDB header: oxidoreductaseChain: B: PDB Molecule: thioredoxin reductase 1;PDBTitle: crystal structure of human thioredoxin reductase 1
93 c3nyeA_
not modelled
19.7
19
PDB header: oxidoreductaseChain: A: PDB Molecule: d-arginine dehydrogenase;PDBTitle: crystal structure of pseudomonas aeruginosa d-arginine dehydrogenase2 in complex with imino-arginine
94 d1dxla1
not modelled
19.5
32
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: FAD/NAD-linked reductases, N-terminal and central domains
95 c3uteB_
not modelled
19.4
35
PDB header: isomeraseChain: B: PDB Molecule: udp-galactopyranose mutase;PDBTitle: crystal structure of aspergillus fumigatus udp galactopyranose mutase2 sulfate complex
96 c1z47B_
not modelled
19.3
29
PDB header: ligand binding proteinChain: B: PDB Molecule: putative abc-transporter atp-binding protein;PDBTitle: structure of the atpase subunit cysa of the putative sulfate atp-2 binding cassette (abc) transporter from alicyclobacillus3 acidocaldarius
97 c2e1mA_
not modelled
19.3
21
PDB header: oxidoreductaseChain: A: PDB Molecule: l-glutamate oxidase;PDBTitle: crystal structure of l-glutamate oxidase from streptomyces sp. x-119-6
98 c4b1bB_
not modelled
18.8
36
PDB header: oxidoreductaseChain: B: PDB Molecule: thioredoxin reductase;PDBTitle: crystal structure of plasmodium falciparum oxidised2 thioredoxin reductase at 2.9 angstrom
99 d3lada1
not modelled
18.8
30
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: FAD/NAD-linked reductases, N-terminal and central domains