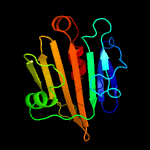
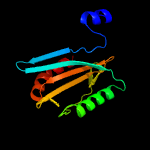
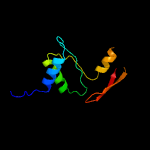
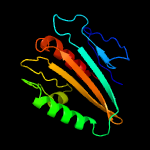
1 c2xb3A_
97.2
14
PDB header: photosynthesisChain: A: PDB Molecule: psbp protein;PDBTitle: the structure of cyanobacterial psbp
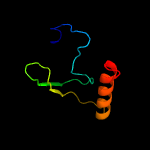
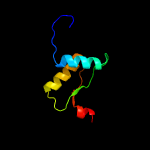
2 c2lnjA_
96.4
12
PDB header: photosynthesisChain: A: PDB Molecule: putative uncharacterized protein sll1418;PDBTitle: solution structure of cyanobacterial psbp (cyanop) from synechocystis2 sp. pcc 6803
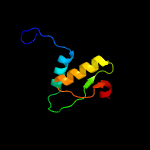
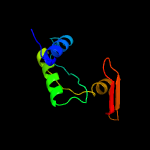
3 d1v2ba_
95.9
14
Fold: Mog1p/PsbP-likeSuperfamily: Mog1p/PsbP-likeFamily: PsbP-like
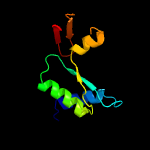
4 d1jcea1
94.8
13
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Actin/HSP70
5 c4ol4A_
94.6
19
PDB header: lipid binding proteinChain: A: PDB Molecule: proline-rich 28 kda antigen;PDBTitle: crystal structure of secreted proline rich antigen mtc28 (rv0040c)2 from mycobacterium tuberculosis
6 d1dkgd1
94.3
14
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Actin/HSP70
7 c2vu4A_
93.5
13
PDB header: photosynthesisChain: A: PDB Molecule: oxygen-evolving enhancer protein 2;PDBTitle: structure of psbp protein from spinacia oleracea at 1.98 a2 resolution
8 c6e8aA_
92.6
25
PDB header: unknown functionChain: A: PDB Molecule: duf1795 domain-containing protein;PDBTitle: crystal structure of dcrb from salmonella enterica at 1.92 angstroms2 resolution
9 d2e8aa1
92.6
10
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Actin/HSP70
10 c2v7yA_
92.5
14
PDB header: chaperoneChain: A: PDB Molecule: chaperone protein dnak;PDBTitle: crystal structure of the molecular chaperone dnak from2 geobacillus kaustophilus hta426 in post-atp hydrolysis3 state
11 d1tu1a_
91.4
13
Fold: Mog1p/PsbP-likeSuperfamily: Mog1p/PsbP-likeFamily: PA0094-like
12 d1bupa1
91.3
11
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Actin/HSP70
13 c4j8fA_
87.0
9
PDB header: chaperoneChain: A: PDB Molecule: heat shock 70 kda protein 1a/1b, hsc70-interacting protein;PDBTitle: crystal structure of a fusion protein containing the nbd of hsp70 and2 the middle domain of hip
14 c5obuA_
85.0
12
PDB header: chaperoneChain: A: PDB Molecule: chaperone protein dnak;PDBTitle: mycoplasma genitalium dnak deletion mutant lacking sbdalpha in complex2 with amppnp.
15 c2khoA_
83.8
13
PDB header: chaperoneChain: A: PDB Molecule: heat shock protein 70;PDBTitle: nmr-rdc / xray structure of e. coli hsp70 (dnak) chaperone (1-605)2 complexed with adp and substrate
16 c3d2fC_
83.1
11
PDB header: chaperoneChain: C: PDB Molecule: heat shock protein homolog sse1;PDBTitle: crystal structure of a complex of sse1p and hsp70
17 c4rtfD_
82.3
16
PDB header: chaperoneChain: D: PDB Molecule: chaperone protein dnak;PDBTitle: crystal structure of molecular chaperone dnak from mycobacterium2 tuberculosis h37rv
18 c3h1qB_
81.5
12
PDB header: structural proteinChain: B: PDB Molecule: ethanolamine utilization protein eutj;PDBTitle: crystal structure of ethanolamine utilization protein eutj from2 carboxydothermus hydrogenoformans
19 c5mb9B_
77.2
16
PDB header: chaperoneChain: B: PDB Molecule: putative heat shock protein;PDBTitle: crystal structure of the eukaryotic ribosome associated complex (rac),2 a unique hsp70/hsp40 pair
20 c3lydA_
73.6
14
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of putative uncharacterized protein from jonesia2 denitrificans
21 c1jcgA_
not modelled
72.4
15
PDB header: structural proteinChain: A: PDB Molecule: rod shape-determining protein mreb;PDBTitle: mreb from thermotoga maritima, amppnp
22 c3c7nB_
not modelled
68.9
11
PDB header: chaperone/chaperoneChain: B: PDB Molecule: heat shock cognate;PDBTitle: structure of the hsp110:hsc70 nucleotide exchange complex
23 c4gniA_
not modelled
68.0
16
PDB header: chaperoneChain: A: PDB Molecule: putative heat shock protein;PDBTitle: structure of the ssz1 atpase bound to atp and magnesium
24 c5e84B_
not modelled
67.6
12
PDB header: chaperoneChain: B: PDB Molecule: 78 kda glucose-regulated protein;PDBTitle: atp-bound state of bip
25 c5jygA_
not modelled
65.0
16
PDB header: structural proteinChain: A: PDB Molecule: actin-like atpase;PDBTitle: cryo-em structure of the mamk filament at 6.5 a
26 c5tkyA_
not modelled
49.6
9
PDB header: chaperoneChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: crystal structure of the co-translational hsp70 chaperone ssb in the2 atp-bound, open conformation
27 c1dkgD_
not modelled
33.4
14
PDB header: complex (hsp24/hsp70)Chain: D: PDB Molecule: molecular chaperone dnak;PDBTitle: crystal structure of the nucleotide exchange factor grpe2 bound to the atpase domain of the molecular chaperone dnak
28 c4pl7B_
not modelled
29.8
13
PDB header: structural protein, contractile proteinChain: B: PDB Molecule: actin,thymosin beta-4;PDBTitle: structure of komagataella pastoris actin-thymosin beta4 hybrid
29 c6gfaA_
not modelled
29.0
16
PDB header: chaperoneChain: A: PDB Molecule: heat shock protein 105 kda;PDBTitle: structure of nucleotide binding domain of hsp110, atp and mg2+2 complexed
30 c3iucC_
not modelled
28.4
11
PDB header: chaperoneChain: C: PDB Molecule: heat shock 70kda protein 5 (glucose-regulatedPDBTitle: crystal structure of the human 70kda heat shock protein 52 (bip/grp78) atpase domain in complex with adp
31 c2lf0A_
not modelled
20.6
55
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein yibl;PDBTitle: solution structure of sf3636, a two-domain unknown function protein2 from shigella flexneri 2a, determined by joint refinement of nmr,3 residual dipolar couplings and small-angle x-ray scattering, nesg4 target sfr339/ocsp target sf3636
32 c5afuB_
not modelled
15.8
15
PDB header: motor proteinChain: B: PDB Molecule: dynactin;PDBTitle: cryo-em structure of dynein tail-dynactin-bicd2n complex
33 c6igmG_
not modelled
15.8
9
PDB header: transcriptionChain: G: PDB Molecule: actin-related protein 6;PDBTitle: cryo-em structure of human srcap complex
34 c5tt2D_
not modelled
15.0
38
PDB header: lyaseChain: D: PDB Molecule: cystathionine gamma-lyase;PDBTitle: inactive conformation of engineered human cystathionine gamma lyase2 (e59n, r119l, e339v) to depleting methionine
35 c4czeA_
not modelled
14.5
14
PDB header: structural proteinChain: A: PDB Molecule: rod shape-determining protein mreb;PDBTitle: c. crescentus mreb, double filament, empty
36 c1o1f4_
not modelled
13.6
13
PDB header: contractile proteinChain: 4: PDB Molecule: skeletal muscle actin;PDBTitle: molecular models of averaged rigor crossbridges from tomograms of2 insect flight muscle
37 c6e5fA_
not modelled
12.8
19
PDB header: lipid binding proteinChain: A: PDB Molecule: lipid binding protein lpqn;PDBTitle: crystal structure of lpqn involved in cell envelope biogenesis of2 mycobacterium tuberculosis
38 c5yk2A_
not modelled
12.4
14
PDB header: transport proteinChain: A: PDB Molecule: probable conserved atp-binding protein abc transporter;PDBTitle: the complex structure of rv3197-erythromycin from mycobacterium2 tuberculosis
39 c4jd2B_
not modelled
10.9
15
PDB header: structural proteinChain: B: PDB Molecule: actin-related protein 2;PDBTitle: crystal structure of bos taurus arp2/3 complex binding with mus2 musculus gmf
40 d1n8pa_
not modelled
10.8
32
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: Cystathionine synthase-like
41 c4pedA_
not modelled
10.6
12
PDB header: transferaseChain: A: PDB Molecule: chaperone activity of bc1 complex-like, mitochondrial;PDBTitle: mitochondrial adck3 employs an atypical protein kinase-like fold to2 enable coenzyme q biosynthes
42 c2nmpC_
not modelled
9.6
37
PDB header: lyaseChain: C: PDB Molecule: cystathionine gamma-lyase;PDBTitle: crystal structure of human cystathionine gamma lyase
43 c2bsjB_
not modelled
9.6
25
PDB header: chaperoneChain: B: PDB Molecule: chaperone protein syct;PDBTitle: native crystal structure of the type iii secretion chaperone syct from2 yersinia enterocolitica
44 d1w96a1
not modelled
9.5
20
Fold: Barrel-sandwich hybridSuperfamily: Rudiment single hybrid motifFamily: BC C-terminal domain-like
45 c3u7iB_
not modelled
9.4
21
PDB header: oxidoreductaseChain: B: PDB Molecule: fmn-dependent nadh-azoreductase 1;PDBTitle: the crystal structure of fmn-dependent nadh-azoreductase 1 (gbaa0966)2 from bacillus anthracis str. ames ancestor
46 c4mtkD_
not modelled
9.0
19
PDB header: toxinChain: D: PDB Molecule: vgrg1;PDBTitle: crystal structure of pa0091 vgrg1, the central spike of the type vi2 secretion system
47 c3lcmB_
not modelled
8.9
36
PDB header: oxidoreductaseChain: B: PDB Molecule: putative oxidoreductase;PDBTitle: crystal structure of smu.1420 from streptococcus mutans ua159
48 d2p5zx1
not modelled
8.9
19
Fold: OB-foldSuperfamily: gp5 N-terminal domain-likeFamily: gp4 N-terminal domain-like
49 c6aufB_
not modelled
8.3
24
PDB header: hydrolaseChain: B: PDB Molecule: beta-lactamase-like protein;PDBTitle: crystal structure of metalo beta lactamases mim-1 from novosphingobium2 pentaromativorans
50 c4kboA_
not modelled
8.3
10
PDB header: signaling proteinChain: A: PDB Molecule: stress-70 protein, mitochondrial;PDBTitle: crystal structure of the human mortalin (grp75) atpase domain in the2 apo form
51 c2khrA_
not modelled
7.7
69
PDB header: biosynthetic proteinChain: A: PDB Molecule: protein mbth;PDBTitle: solution structure of rv2377c, a mbth-like protein from mycobacterium2 tuberculosis
52 c5jn6A_
not modelled
7.6
22
PDB header: unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: the nmr solution structure of rpa3313
53 d1w96c1
not modelled
7.5
20
Fold: Barrel-sandwich hybridSuperfamily: Rudiment single hybrid motifFamily: BC C-terminal domain-like
54 c6e0sA_
not modelled
7.5
15
PDB header: hydrolaseChain: A: PDB Molecule: mem-a1;PDBTitle: crystal structure of mem-a1, a subclass b3 metallo-beta-lactamase2 isolated from a soil metagenome library
55 c2v9cA_
not modelled
7.4
21
PDB header: oxidoreductaseChain: A: PDB Molecule: fmn-dependent nadh-azoreductase 1;PDBTitle: x-ray crystallographic structure of a pseudomonas2 aeruginosa azoreductase in complex with methyl red.
56 d1ig4a_
not modelled
7.1
20
Fold: DNA-binding domainSuperfamily: DNA-binding domainFamily: Methyl-CpG-binding domain, MBD
57 c2lpdA_
not modelled
7.0
36
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: solution structure of a mbth-like protein from burkholderia2 pseudomallei, the etiological agent responsible for melioidosis,3 seattle structural genomics center for infectious disease target4 bupsa.13472.b
58 c5u89B_
not modelled
7.0
23
PDB header: hydrolase/inhibitorChain: B: PDB Molecule: mbth domain protein;PDBTitle: crystal structure of a cross-module fragment from the dimodular nrps2 dhbf
59 d1ei5a2
not modelled
6.8
31
Fold: Streptavidin-likeSuperfamily: D-aminopeptidase, middle and C-terminal domainsFamily: D-aminopeptidase, middle and C-terminal domains
60 d1dy2a_
not modelled
6.6
27
Fold: C-type lectin-likeSuperfamily: C-type lectin-likeFamily: Endostatin
61 c5oqml_
not modelled
6.6
22
PDB header: transcriptionChain: L: PDB Molecule: dna-directed rna polymerases i, ii, and iii subunit rpabc4;PDBTitle: structure of yeast transcription pre-initiation complex with tfiih and2 core mediator
62 d1hc8a_
not modelled
6.4
6
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Ribosomal protein L11, C-terminal domainFamily: Ribosomal protein L11, C-terminal domain
63 d1mmsa1
not modelled
6.4
12
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Ribosomal protein L11, C-terminal domainFamily: Ribosomal protein L11, C-terminal domain
64 c2ky8A_
not modelled
6.3
24
PDB header: transcription/dnaChain: A: PDB Molecule: methyl-cpg-binding domain protein 2;PDBTitle: solution structure and dynamic analysis of chicken mbd2 methyl binding2 domain bound to a target methylated dna sequence
65 d1xbpg1
not modelled
6.2
6
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Ribosomal protein L11, C-terminal domainFamily: Ribosomal protein L11, C-terminal domain
66 d1rlia_
not modelled
6.2
15
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Hypothetical protein YwqN
67 c5f4bB_
not modelled
6.2
8
PDB header: oxidoreductaseChain: B: PDB Molecule: nad(p)h dehydrogenase (quinone);PDBTitle: structure of b. abortus wrba-related protein a (wrpa)
68 c3rpeA_
not modelled
6.1
29
PDB header: oxidoreductaseChain: A: PDB Molecule: modulator of drug activity b;PDBTitle: 1.1 angstrom crystal structure of putative modulator of drug activity2 (mdab) from yersinia pestis co92.
69 d2gycg1
not modelled
5.7
6
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Ribosomal protein L11, C-terminal domainFamily: Ribosomal protein L11, C-terminal domain
70 d1vqoi1
not modelled
5.5
18
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Ribosomal protein L11, C-terminal domainFamily: Ribosomal protein L11, C-terminal domain
71 c1vq8I_
not modelled
5.5
18
PDB header: ribosomeChain: I: PDB Molecule: 50s ribosomal protein l11p;PDBTitle: the structure of ccda-phe-cap-bio and the antibiotic sparsomycin bound2 to the large ribosomal subunit of haloarcula marismortui
72 d1d4aa_
not modelled
5.5
21
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Quinone reductase
73 c5iqkB_
not modelled
5.5
14
PDB header: hydrolaseChain: B: PDB Molecule: beta-lactamase rm3;PDBTitle: rm3 metallo-beta-lactamase
74 d2qwxa1
not modelled
5.3
21
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Quinone reductase
75 c4r81C_
not modelled
5.2
36
PDB header: oxidoreductaseChain: C: PDB Molecule: nadh dehydrogenase;PDBTitle: nad(p)h:quinone oxidoreductase from methanothermobacter marburgensis
76 c2zkiH_
not modelled
5.2
21
PDB header: transcriptionChain: H: PDB Molecule: 199aa long hypothetical trp repressor bindingPDBTitle: crystal structure of hypothetical trp repressor binding2 protein from sul folobus tokodaii (st0872)
77 c5o5oD_
not modelled
5.1
31
PDB header: chaperoneChain: D: PDB Molecule: rnase adapter protein rapz;PDBTitle: x-ray crystal structure of rapz from escherichia coli (p32 space2 group)
78 d1c3ga1
not modelled
5.1
2
Fold: HSP40/DnaJ peptide-binding domainSuperfamily: HSP40/DnaJ peptide-binding domainFamily: HSP40/DnaJ peptide-binding domain
79 c2lazA_
not modelled
5.1
21
PDB header: signaling protein/transcriptionChain: A: PDB Molecule: e3 ubiquitin-protein ligase smurf1;PDBTitle: structure of the first ww domain of human smurf1 in complex with a2 mono-phosphorylated human smad1 derived peptide