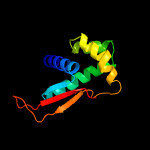
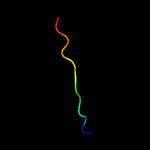1 c5o60O_
100.0
92
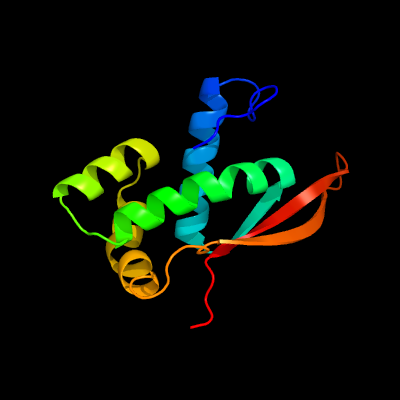
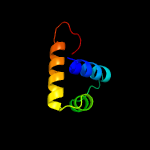
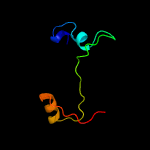
PDB header: ribosomeChain: O: PDB Molecule: 50s ribosomal protein l17;PDBTitle: structure of the 50s large ribosomal subunit from mycobacterium2 smegmatis
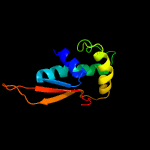
2 c1vw4L_
100.0
30
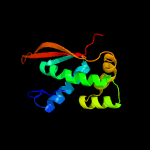
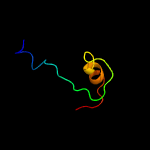
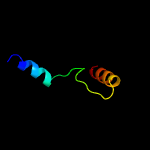
PDB header: ribosomeChain: L: PDB Molecule: 54s ribosomal protein l8, mitochondrial;PDBTitle: structure of the yeast mitochondrial large ribosomal subunit
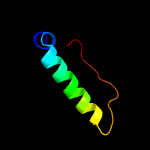
3 d2qamn1
100.0
49
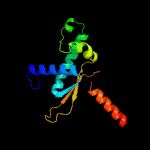
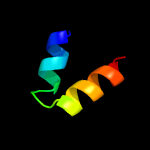
Fold: Prokaryotic ribosomal protein L17Superfamily: Prokaryotic ribosomal protein L17Family: Prokaryotic ribosomal protein L17
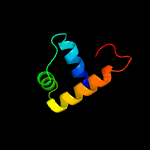
4 c4v19R_
100.0
37
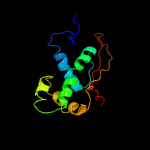
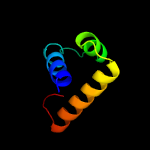
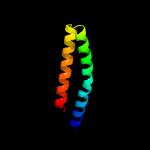
PDB header: ribosomeChain: R: PDB Molecule: mitoribosomal protein bl17m, mrpl17;PDBTitle: structure of the large subunit of the mammalian mitoribosome, part 12 of 2
5 c3bboP_
100.0
50
PDB header: ribosomeChain: P: PDB Molecule: ribosomal protein l17;PDBTitle: homology model for the spinach chloroplast 50s subunit fitted to 9.4a2 cryo-em map of the 70s chlororibosome
6 d2zjrk1
100.0
47
Fold: Prokaryotic ribosomal protein L17Superfamily: Prokaryotic ribosomal protein L17Family: Prokaryotic ribosomal protein L17
7 c3j3vN_
100.0
46
PDB header: ribosomeChain: N: PDB Molecule: 50s ribosomal protein l17;PDBTitle: atomic model of the immature 50s subunit from bacillus subtilis (state2 i-a)
8 d1gd8a_
100.0
54
Fold: Prokaryotic ribosomal protein L17Superfamily: Prokaryotic ribosomal protein L17Family: Prokaryotic ribosomal protein L17
9 d2cqma1
100.0
40
Fold: Prokaryotic ribosomal protein L17Superfamily: Prokaryotic ribosomal protein L17Family: Prokaryotic ribosomal protein L17
10 c5vn4A_
36.5
13
PDB header: transferaseChain: A: PDB Molecule: adenine phosphoribosyltransferase, putative;PDBTitle: crystal structure of adenine phosphoribosyl transferase from2 trypanosoma brucei in complex with amp, pyrophosphate, and ribose-5-3 phosphate
11 c3w6kC_
35.8
11
PDB header: cell cycleChain: C: PDB Molecule: scpb;PDBTitle: crystal structure of dimer of scpb n-terminal domain complexed with2 scpa peptide
12 c3w6jC_
33.8
11
PDB header: cell cycleChain: C: PDB Molecule: scpb;PDBTitle: crystal structure of scpab core complex
13 c5yhhA_
33.5
22
PDB header: oxidoreductaseChain: A: PDB Molecule: uncharacterized conserved protein yiim;PDBTitle: crystal structure of yiim from geobacillus stearothermophilus
14 c3h0dB_
24.6
50
PDB header: transcription/dnaChain: B: PDB Molecule: ctsr;PDBTitle: crystal structure of ctsr in complex with a 26bp dna duplex
15 c2z99A_
21.1
13
PDB header: cell cycleChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: crystal structure of scpb from mycobacterium tuberculosis
16 d1o65a_
20.3
16
Fold: PK beta-barrel domain-likeSuperfamily: PK beta-barrel domain-likeFamily: MOSC (MOCO sulphurase C-terminal) domain
17 d1xd7a_
16.5
11
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Transcriptional regulator Rrf2
18 c3kowH_
14.8
22
PDB header: metal binding proteinChain: H: PDB Molecule: d-ornithine aminomutase s component;PDBTitle: crystal structure of ornithine 4,5 aminomutase backsoaked complex
19 c5he9E_
14.1
33
PDB header: protein bindingChain: E: PDB Molecule: phage inhibitor protein;PDBTitle: bacterial initiation protein in complex with phage inhibitor protein
20 c2ic6B_
13.4
21
PDB header: viral proteinChain: B: PDB Molecule: nucleocapsid protein;PDBTitle: the coiled-coil domain (residues 1-75) structure of the sin nombre2 virus nucleocapsid protein
21 c5wlbF_
not modelled
11.5
71
PDB header: protein bindingChain: F: PDB Molecule: 225-15 b;PDBTitle: kras g12v, bound to gppnhp and miniprotein 225-15a/b
22 d1v66a_
not modelled
11.3
23
Fold: LEM/SAP HeH motifSuperfamily: SAP domainFamily: SAP domain
23 c2op8A_
not modelled
11.2
10
PDB header: isomeraseChain: A: PDB Molecule: probable tautomerase ywhb;PDBTitle: crystal structure of ywhb- homologue of 4-oxalocrotonate tautomerase
24 c5tv6A_
not modelled
10.7
19
PDB header: ligaseChain: A: PDB Molecule: 6-carboxyhexanoate--coa ligase;PDBTitle: a. aeolicus biow with pimelate
25 c1x4qA_
not modelled
10.4
29
PDB header: rna binding proteinChain: A: PDB Molecule: u4/u6 small nuclear ribonucleoprotein prp3;PDBTitle: solution structure of pwi domain in u4/u6 small nuclear2 ribonucleoprotein prp3(hprp3)
26 d1t6sa1
not modelled
10.1
10
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: ScpB/YpuH-like
27 c5wlbC_
not modelled
9.5
71
PDB header: protein bindingChain: C: PDB Molecule: 225-15 b;PDBTitle: kras g12v, bound to gppnhp and miniprotein 225-15a/b
28 c4fipG_
not modelled
9.0
55
PDB header: hydrolaseChain: G: PDB Molecule: saga-associated factor 11;PDBTitle: structure of the saga ubp8(s144n)/sgf11(1-72, delta-znf)/sus1/sgf732 dub module
29 c4dtfA_
not modelled
8.5
24
PDB header: toxinChain: A: PDB Molecule: vgrg protein;PDBTitle: structure of a vgrg vibrio cholerae toxin acd domain in complex with2 amp-pnp and mg++
30 c2wb1J_
not modelled
8.3
25
PDB header: transcriptionChain: J: PDB Molecule: dna-directed rna polymerase rpo13 subunit;PDBTitle: the complete structure of the archaeal 13-subunit dna-2 directed rna polymerase
31 c1pqrA_
not modelled
7.7
44
PDB header: toxinChain: A: PDB Molecule: alpha-a-conotoxin eiva;PDBTitle: solution conformation of alphaa-conotoxin eiva
32 d1otfa_
not modelled
7.3
5
Fold: Tautomerase/MIFSuperfamily: Tautomerase/MIFFamily: 4-oxalocrotonate tautomerase-like
33 c2llrA_
not modelled
7.0
45
PDB header: antimicrobial proteinChain: A: PDB Molecule: alvinellacin;PDBTitle: nmr structure of alvinellacin
34 c4i0wA_
not modelled
7.0
10
PDB header: hydrolaseChain: A: PDB Molecule: protease cspb;PDBTitle: structure of the clostridium perfringens cspb protease
35 c3ry0A_
not modelled
6.9
24
PDB header: isomeraseChain: A: PDB Molecule: putative tautomerase;PDBTitle: crystal structure of tomn, a 4-oxalocrotonate tautomerase homologue in2 tomaymycin biosynthetic pathway
36 c2bnlE_
not modelled
6.8
7
PDB header: stress-responseChain: E: PDB Molecule: modulator protein rsbr;PDBTitle: the structure of the n-terminal domain of rsbr
37 c4acrA_
not modelled
6.3
9
PDB header: membrane proteinChain: A: PDB Molecule: glypican-1;PDBTitle: crystal structure of n-glycosylated, c-terminally truncated human2 glypican-1
38 d1qb7a_
not modelled
6.3
13
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)
39 c2cqjA_
not modelled
6.3
21
PDB header: rna binding proteinChain: A: PDB Molecule: u3 small nucleolar ribonucleoprotein proteinPDBTitle: solution structure of the s4 domain of u3 small nucleolar2 ribonucleoprotein protein imp3 homolog
40 c4fdxB_
not modelled
6.2
10
PDB header: isomeraseChain: B: PDB Molecule: 4-oxalocrotonase tautomerase isozyme;PDBTitle: kinetic and structural characterization of the 4-oxalocrotonate2 tautomerase isozymes from methylibium petroleiphilum
41 d1bjpa_
not modelled
6.1
10
Fold: Tautomerase/MIFSuperfamily: Tautomerase/MIFFamily: 4-oxalocrotonate tautomerase-like
42 c4if2A_
not modelled
6.1
26
PDB header: hydrolaseChain: A: PDB Molecule: phosphotriesterase homology protein;PDBTitle: structure of the phosphotriesterase from mycobacterium tuberculosis
43 c2x4kB_
not modelled
5.8
14
PDB header: isomeraseChain: B: PDB Molecule: 4-oxalocrotonate tautomerase;PDBTitle: crystal structure of sar1376, a putative 4-oxalocrotonate2 tautomerase from the methicillin-resistant staphylococcus3 aureus (mrsa)
44 c5x5wC_
not modelled
5.6
27
PDB header: viral protein/cell adhesionChain: C: PDB Molecule: gd;PDBTitle: crystal structure of pseudorabies virus glycoprotein d
45 c3mkoA_
not modelled
5.6
9
PDB header: viral proteinChain: A: PDB Molecule: glycoprotein c;PDBTitle: crystal structure of the lymphocytic choriomeningitis virus membrane2 fusion glycoprotein gp2 in its postfusion conformation
46 c5wd9A_
not modelled
5.6
21
PDB header: protein bindingChain: A: PDB Molecule: lem22;PDBTitle: crystal structure of legionella pneumophila effector lpg2328
47 d2incb1
not modelled
5.5
15
Fold: Ferritin-likeSuperfamily: Ferritin-likeFamily: Ribonucleotide reductase-like
48 c6nd4K_
not modelled
5.4
20
PDB header: ribosomeChain: K: PDB Molecule: utp9;PDBTitle: conformational switches control early maturation of the eukaryotic2 small ribosomal subunit
49 d1ht6a1
not modelled
5.3
17
Fold: Glycosyl hydrolase domainSuperfamily: Glycosyl hydrolase domainFamily: alpha-Amylases, C-terminal beta-sheet domain
50 c2d3o1_
not modelled
5.2
54
PDB header: ribosomeChain: 1: PDB Molecule: trigger factor;PDBTitle: structure of ribosome binding domain of the trigger factor on the 50s2 ribosomal subunit from d. radiodurans
51 c3kmiB_
not modelled
5.1
10
PDB header: membrane proteinChain: B: PDB Molecule: putative membrane protein cog4129;PDBTitle: crystal structure of putative membrane protein from clostridium2 difficile 630
52 c3dhgB_
not modelled
5.1
21
PDB header: oxidoreductaseChain: B: PDB Molecule: toluene 4-monooxygenase hydroxylase beta subunit;PDBTitle: crystal structure of toluene 4-monoxygenase hydroxylase