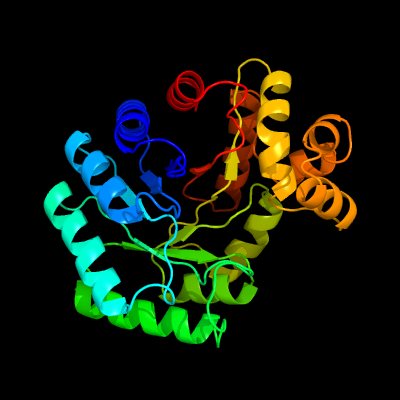
| 1 | c1z69D_
|
|
 |
100.0 |
17 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:coenzyme f420-dependent n(5),n(10)-
PDBTitle: crystal structure of methylenetetrahydromethanopterin2 reductase (mer) in complex with coenzyme f420
|
|
|
|
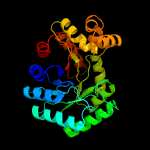
| 2 | d1ezwa_
|
|
 |
100.0 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Bacterial luciferase-like
Family:F420 dependent oxidoreductases |
|
|
|
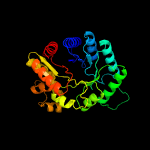
| 3 | d1rhca_
|
|
 |
100.0 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Bacterial luciferase-like
Family:F420 dependent oxidoreductases |
|
|
|
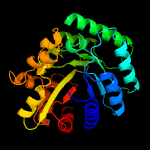
| 4 | d1f07a_
|
|
 |
100.0 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Bacterial luciferase-like
Family:F420 dependent oxidoreductases |
|
|
|
| 5 | d1nqka_
|
|
 |
100.0 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Bacterial luciferase-like
Family:Ssud-like monoxygenases |
|
|
|
| 6 | c3sdoB_
|
|
 |
100.0 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:nitrilotriacetate monooxygenase;
PDBTitle: structure of a nitrilotriacetate monooxygenase from burkholderia2 pseudomallei
|
|
|
|
| 7 | c3raoB_
|
|
 |
100.0 |
17 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:putative luciferase-like monooxygenase;
PDBTitle: crystal structure of the luciferase-like monooxygenase from bacillus2 cereus atcc 10987.
|
|
|
|
| 8 | c5tlcA_
|
|
 |
100.0 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dibenzothiophene desulfurization enzyme a;
PDBTitle: crystal structure of bdsa from bacillus subtilis wu-s2b
|
|
|
|
| 9 | c3c8nB_
|
|
 |
100.0 |
20 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:probable f420-dependent glucose-6-phosphate dehydrogenase
PDBTitle: crystal structure of apo-fgd1 from mycobacterium tuberculosis
|
|
|
|
| 10 | c2i7gA_
|
|
 |
100.0 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:monooxygenase;
PDBTitle: crystal structure of monooxygenase from agrobacterium tumefaciens
|
|
|
|
| 11 | d1luca_
|
|
 |
100.0 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Bacterial luciferase-like
Family:Bacterial luciferase (alkanal monooxygenase) |
|
|
|
| 12 | d1lucb_
|
|
 |
100.0 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Bacterial luciferase-like
Family:Bacterial luciferase (alkanal monooxygenase) |
|
|
|
| 13 | c5w4zA_
|
|
 |
100.0 |
16 |
PDB header:flavoprotein
Chain: A: PDB Molecule:riboflavin lyase;
PDBTitle: crystal structure of riboflavin lyase (rcae) with modified fmn and2 substrate riboflavin
|
|
|
|
| 14 | c1tvlA_
|
|
 |
100.0 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:protein ytnj;
PDBTitle: structure of ytnj from bacillus subtilis
|
|
|
|
| 15 | d1tvla_
|
|
 |
100.0 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Bacterial luciferase-like
Family:Ssud-like monoxygenases |
|
|
|
| 16 | c2wgkA_
|
|
 |
100.0 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:3,6-diketocamphane 1,6 monooxygenase;
PDBTitle: type ii baeyer-villiger monooxygenase oxygenating2 constituent of 3,6-diketocamphane 1,6 monooxygenase from3 pseudomonas putida
|
|
|
|
| 17 | c2b81D_
|
|
 |
100.0 |
17 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:luciferase-like monooxygenase;
PDBTitle: crystal structure of the luciferase-like monooxygenase from bacillus2 cereus
|
|
|
|
| 18 | c5dqpA_
|
|
 |
100.0 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:edta monooxygenase;
PDBTitle: edta monooxygenase (emoa) from chelativorans sp. bnc1
|
|
|
|
| 19 | c3b9nB_
|
|
 |
100.0 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:alkane monoxygenase;
PDBTitle: crystal structure of long-chain alkane monooxygenase (lada)
|
|
|
|
| 20 | c5wanA_
|
|
 |
100.0 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:pyrimidine monooxygenase ruta;
PDBTitle: crystal structure of a flavoenzyme ruta in the pyrimidine catabolic2 pathway
|
|
|
|
| 21 | c6friD_ |
|
not modelled |
100.0 |
14 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:alkanal monooxygenase beta chain;
PDBTitle: structure of luxb from photobacterium leiognathi
|
|
|
| 22 | c6ak1B_ |
|
not modelled |
100.0 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:dimethyl-sulfide monooxygenase;
PDBTitle: crystal structure of dmoa from hyphomicrobium sulfonivorans
|
|
|
| 23 | d1nfpa_ |
|
not modelled |
99.5 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Bacterial luciferase-like
Family:Non-fluorescent flavoprotein (luxF, FP390) |
|
|
| 24 | d1fvpa_ |
|
not modelled |
98.0 |
9 |
Fold:TIM beta/alpha-barrel
Superfamily:Bacterial luciferase-like
Family:Non-fluorescent flavoprotein (luxF, FP390) |
|
|
| 25 | d2d69a1 |
|
not modelled |
89.0 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:RuBisCo, C-terminal domain
Family:RuBisCo, large subunit, C-terminal domain |
|
|
| 26 | c6hunA_ |
|
not modelled |
88.0 |
18 |
PDB header:photosynthesis
Chain: A: PDB Molecule:ribulose bisphosphate carboxylase;
PDBTitle: dimeric archeal rubisco from hyperthermus butylicus
|
|
|
| 27 | c2d69B_ |
|
not modelled |
86.4 |
16 |
PDB header:lyase
Chain: B: PDB Molecule:ribulose bisphosphate carboxylase;
PDBTitle: crystal structure of the complex of sulfate ion and octameric2 ribulose-1,5-bisphosphate carboxylase/oxygenase (rubisco) from3 pyrococcus horikoshii ot3 (form-2 crystal)
|
|
|
| 28 | d1geha1 |
|
not modelled |
86.3 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:RuBisCo, C-terminal domain
Family:RuBisCo, large subunit, C-terminal domain |
|
|
| 29 | d8ruca1 |
|
not modelled |
83.2 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:RuBisCo, C-terminal domain
Family:RuBisCo, large subunit, C-terminal domain |
|
|
| 30 | c1gehE_ |
|
not modelled |
82.6 |
16 |
PDB header:lyase
Chain: E: PDB Molecule:ribulose-1,5-bisphosphate carboxylase/oxygenase;
PDBTitle: crystal structure of archaeal rubisco (ribulose 1,5-bisphosphate2 carboxylase/oxygenase)
|
|
|
| 31 | c1rcxH_ |
|
not modelled |
81.2 |
20 |
PDB header:lyase (carbon-carbon)
Chain: H: PDB Molecule:ribulose bisphosphate carboxylase/oxygenase;
PDBTitle: non-activated spinach rubisco in complex with its substrate2 ribulose-1,5-bisphosphate
|
|
|
| 32 | d1wdda1 |
|
not modelled |
78.4 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:RuBisCo, C-terminal domain
Family:RuBisCo, large subunit, C-terminal domain |
|
|
| 33 | d1tzza1 |
|
not modelled |
76.2 |
9 |
Fold:TIM beta/alpha-barrel
Superfamily:Enolase C-terminal domain-like
Family:D-glucarate dehydratase-like |
|
|
| 34 | d1rbla1 |
|
not modelled |
75.1 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:RuBisCo, C-terminal domain
Family:RuBisCo, large subunit, C-terminal domain |
|
|
| 35 | c1rldB_ |
|
not modelled |
74.8 |
20 |
PDB header:lyase(carbon-carbon)
Chain: B: PDB Molecule:ribulose 1,5 bisphosphate carboxylase/oxygenase (large
PDBTitle: solid-state phase transition in the crystal structure of ribulose 1,5-2 biphosphate carboxylase(slash)oxygenase
|
|
|
| 36 | d1s2wa_ |
|
not modelled |
74.4 |
23 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Phosphoenolpyruvate mutase/Isocitrate lyase-like |
|
|
| 37 | d1ej7l1 |
|
not modelled |
70.2 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:RuBisCo, C-terminal domain
Family:RuBisCo, large subunit, C-terminal domain |
|
|
| 38 | d1jpdx1 |
|
not modelled |
69.6 |
23 |
Fold:TIM beta/alpha-barrel
Superfamily:Enolase C-terminal domain-like
Family:D-glucarate dehydratase-like |
|
|
| 39 | c4ggbA_ |
|
not modelled |
68.4 |
12 |
PDB header:isomerase
Chain: A: PDB Molecule:putative uncharacterized protein;
PDBTitle: crystal structure of a proposed galactarolactone cycloisomerase from2 agrobacterium tumefaciens, target efi-500704, with bound ca,3 disordered loops
|
|
|
| 40 | d1svda1 |
|
not modelled |
68.2 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:RuBisCo, C-terminal domain
Family:RuBisCo, large subunit, C-terminal domain |
|
|
| 41 | c3nwrA_ |
|
not modelled |
67.9 |
12 |
PDB header:lyase
Chain: A: PDB Molecule:a rubisco-like protein;
PDBTitle: crystal structure of a rubisco-like protein from burkholderia fungorum
|
|
|
| 42 | c5zmyF_ |
|
not modelled |
64.6 |
21 |
PDB header:hydrolase
Chain: F: PDB Molecule:cis-epoxysuccinate hydrolase;
PDBTitle: crystal structure of a cis-epoxysuccinate hydrolase producing d(-)-2 tartaric acids
|
|
|
| 43 | c3lyeA_ |
|
not modelled |
63.8 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:oxaloacetate acetyl hydrolase;
PDBTitle: crystal structure of oxaloacetate acetylhydrolase
|
|
|
| 44 | d1ykwa1 |
|
not modelled |
63.5 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:RuBisCo, C-terminal domain
Family:RuBisCo, large subunit, C-terminal domain |
|
|
| 45 | c3fa4D_ |
|
not modelled |
63.3 |
23 |
PDB header:lyase
Chain: D: PDB Molecule:2,3-dimethylmalate lyase;
PDBTitle: crystal structure of 2,3-dimethylmalate lyase, a pep mutase/isocitrate2 lyase superfamily member, triclinic crystal form
|
|
|
| 46 | c2qdeA_ |
|
not modelled |
62.9 |
18 |
PDB header:lyase
Chain: A: PDB Molecule:mandelate racemase/muconate lactonizing enzyme family
PDBTitle: crystal structure of mandelate racemase/muconate lactonizing family2 protein from azoarcus sp. ebn1
|
|
|
| 47 | c2nqlB_ |
|
not modelled |
62.9 |
16 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:isomerase/lactonizing enzyme;
PDBTitle: crystal structure of a member of the enolase superfamily from2 agrobacterium tumefaciens
|
|
|
| 48 | c3qfwB_ |
|
not modelled |
61.2 |
17 |
PDB header:lyase
Chain: B: PDB Molecule:ribulose-1,5-bisphosphate carboxylase/oxygenase large
PDBTitle: crystal structure of rubisco-like protein from rhodopseudomonas2 palustris
|
|
|
| 49 | d2gdqa1 |
|
not modelled |
60.0 |
9 |
Fold:TIM beta/alpha-barrel
Superfamily:Enolase C-terminal domain-like
Family:D-glucarate dehydratase-like |
|
|
| 50 | c4jhmC_ |
|
not modelled |
59.7 |
9 |
PDB header:isomerase
Chain: C: PDB Molecule:mandelate racemase / muconate lactonizing enzyme, c-
PDBTitle: crystal structure of a putative mandelate racemase/muconate2 lactonizing enzyme from pseudovibrio sp.
|
|
|
| 51 | c3ddmD_ |
|
not modelled |
58.7 |
13 |
PDB header:lyase
Chain: D: PDB Molecule:putative mandelate racemase/muconate lactonizing
PDBTitle: crystal structure of mandelate racemase/muconate2 lactonizing enzyme from bordetella bronchiseptica rb50
|
|
|
| 52 | c1zlpA_ |
|
not modelled |
57.7 |
22 |
PDB header:lyase
Chain: A: PDB Molecule:petal death protein;
PDBTitle: petal death protein psr132 with cysteine-linked glutaraldehyde forming2 a thiohemiacetal adduct
|
|
|
| 53 | c2hjpA_ |
|
not modelled |
56.9 |
20 |
PDB header:hydrolase
Chain: A: PDB Molecule:phosphonopyruvate hydrolase;
PDBTitle: crystal structure of phosphonopyruvate hydrolase complex with2 phosphonopyruvate and mg++
|
|
|
| 54 | c2qygC_ |
|
not modelled |
56.8 |
14 |
PDB header:unknown function
Chain: C: PDB Molecule:ribulose bisphosphate carboxylase-like protein 2;
PDBTitle: crystal structure of a rubisco-like protein rlp2 from rhodopseudomonas2 palustris
|
|
|
| 55 | c2oemA_ |
|
not modelled |
56.7 |
24 |
PDB header:isomerase
Chain: A: PDB Molecule:2,3-diketo-5-methylthiopentyl-1-phosphate enolase;
PDBTitle: crystal structure of a rubisco-like protein from geobacillus2 kaustophilus liganded with mg2+ and 2,3-diketohexane 1-phosphate
|
|
|
| 56 | c2qddA_ |
|
not modelled |
56.6 |
18 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:mandelate racemase/muconate lactonizing enzyme;
PDBTitle: crystal structure of a member of enolase superfamily from roseovarius2 nubinhibens ism
|
|
|
| 57 | c2ps2A_ |
|
not modelled |
56.6 |
16 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative mandelate racemase/muconate lactonizing enzyme;
PDBTitle: crystal structure of putative mandelate racemase/muconate lactonizing2 enzyme from aspergillus oryzae
|
|
|
| 58 | d2gl5a1 |
|
not modelled |
56.5 |
24 |
Fold:TIM beta/alpha-barrel
Superfamily:Enolase C-terminal domain-like
Family:D-glucarate dehydratase-like |
|
|
| 59 | c2rdxG_ |
|
not modelled |
56.4 |
18 |
PDB header:structural genomics, unknown function
Chain: G: PDB Molecule:mandelate racemase/muconate lactonizing enzyme, putative;
PDBTitle: crystal structure of mandelate racemase/muconate lactonizing enzyme2 from roseovarius nubinhibens ism
|
|
|
| 60 | d1m53a2 |
|
not modelled |
54.9 |
22 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
|
|
| 61 | c3h12B_ |
|
not modelled |
54.9 |
18 |
PDB header:isomerase
Chain: B: PDB Molecule:mandelate racemase;
PDBTitle: crystal structure of putative mandelate racemase from bordetella2 bronchiseptica rb50
|
|
|
| 62 | d1gk8a1 |
|
not modelled |
54.6 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:RuBisCo, C-terminal domain
Family:RuBisCo, large subunit, C-terminal domain |
|
|
| 63 | d1rvka1 |
|
not modelled |
54.4 |
11 |
Fold:TIM beta/alpha-barrel
Superfamily:Enolase C-terminal domain-like
Family:D-glucarate dehydratase-like |
|
|
| 64 | c4nasD_ |
|
not modelled |
53.8 |
19 |
PDB header:lyase
Chain: D: PDB Molecule:ribulose-bisphosphate carboxylase;
PDBTitle: the crystal structure of a rubisco-like protein (mtnw) from2 alicyclobacillus acidocaldarius subsp. acidocaldarius dsm 446
|
|
|
| 65 | c3fk4A_ |
|
not modelled |
53.7 |
21 |
PDB header:isomerase
Chain: A: PDB Molecule:rubisco-like protein;
PDBTitle: crystal structure of rubisco-like protein from bacillus2 cereus atcc 14579
|
|
|
| 66 | c3noeA_ |
|
not modelled |
53.6 |
11 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from pseudomonas2 aeruginosa
|
|
|
| 67 | c3ih1A_ |
|
not modelled |
52.9 |
21 |
PDB header:lyase
Chain: A: PDB Molecule:methylisocitrate lyase;
PDBTitle: crystal structure of carboxyvinyl-carboxyphosphonate phosphorylmutase2 from bacillus anthracis
|
|
|
| 68 | c2zviB_ |
|
not modelled |
52.6 |
19 |
PDB header:isomerase
Chain: B: PDB Molecule:2,3-diketo-5-methylthiopentyl-1-phosphate
PDBTitle: crystal structure of 2,3-diketo-5-methylthiopentyl-1-2 phosphate enolase from bacillus subtilis
|
|
|
| 69 | c1bf2A_ |
|
not modelled |
52.5 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:isoamylase;
PDBTitle: structure of pseudomonas isoamylase
|
|
|
| 70 | d1xxxa1 |
|
not modelled |
51.4 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
|
|
| 71 | c4mggB_ |
|
not modelled |
50.6 |
11 |
PDB header:isomerase
Chain: B: PDB Molecule:muconate lactonizing enzyme;
PDBTitle: crystal structure of an enolase (mandelate racemase subgroup) from2 labrenzia aggregata iam 12614 (target nysgrc-012903) with bound mg,3 space group p212121
|
|
|
| 72 | c3eooL_ |
|
not modelled |
50.5 |
22 |
PDB header:lyase
Chain: L: PDB Molecule:methylisocitrate lyase;
PDBTitle: 2.9a crystal structure of methyl-isocitrate lyase from2 burkholderia pseudomallei
|
|
|
| 73 | c1telA_ |
|
not modelled |
50.4 |
17 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:ribulose bisphosphate carboxylase, large subunit;
PDBTitle: crystal structure of a rubisco-like protein from chlorobium2 tepidum
|
|
|
| 74 | c3daqB_ |
|
not modelled |
50.2 |
18 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from methicillin-2 resistant staphylococcus aureus
|
|
|
| 75 | d1muma_ |
|
not modelled |
50.0 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Phosphoenolpyruvate mutase/Isocitrate lyase-like |
|
|
| 76 | c5uncB_ |
|
not modelled |
49.8 |
23 |
PDB header:isomerase
Chain: B: PDB Molecule:phosphoenolpyruvate phosphomutase;
PDBTitle: the crystal structure of phosphoenolpyruvate phosphomutase from2 streptomyces platensis subsp. rosaceus
|
|
|
| 77 | d1lwha2 |
|
not modelled |
49.4 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
|
|
| 78 | c2qiwA_ |
|
not modelled |
49.1 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:pep phosphonomutase;
PDBTitle: crystal structure of a putative phosphoenolpyruvate phosphonomutase2 (ncgl1015, cgl1060) from corynebacterium glutamicum atcc 13032 at3 1.80 a resolution
|
|
|
| 79 | c4xkyC_ |
|
not modelled |
48.9 |
20 |
PDB header:lyase
Chain: C: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: structure of dihydrodipicolinate synthase from the commensal bacterium2 bacteroides thetaiotaomicron at 2.1 a resolution
|
|
|
| 80 | c4kemB_ |
|
not modelled |
48.4 |
7 |
PDB header:lyase
Chain: B: PDB Molecule:mandelate racemase/muconate lactonizing enzyme;
PDBTitle: crystal structure of a tartrate dehydratase from azospirillum, target2 efi-502395, with bound mg and a putative acrylate ion, ordered active3 site
|
|
|
| 81 | c3ritE_ |
|
not modelled |
48.3 |
18 |
PDB header:isomerase
Chain: E: PDB Molecule:dipeptide epimerase;
PDBTitle: crystal structure of dipeptide epimerase from methylococcus capsulatus2 complexed with mg and dipeptide l-arg-d-lys
|
|
|
| 82 | c3jw7E_ |
|
not modelled |
48.3 |
18 |
PDB header:isomerase
Chain: E: PDB Molecule:dipeptide epimerase;
PDBTitle: crystal structure of dipeptide epimerase from enterococcus faecalis2 v583 complexed with mg and dipeptide l-ile-l-tyr
|
|
|
| 83 | c3bjsB_ |
|
not modelled |
48.0 |
13 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:mandelate racemase/muconate lactonizing enzyme;
PDBTitle: crystal structure of a member of enolase superfamily from polaromonas2 sp. js666
|
|
|
| 84 | c3fluD_ |
|
not modelled |
47.9 |
16 |
PDB header:lyase
Chain: D: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from the pathogen2 neisseria meningitidis
|
|
|
| 85 | c3m07A_ |
|
not modelled |
47.7 |
23 |
PDB header:unknown function
Chain: A: PDB Molecule:putative alpha amylase;
PDBTitle: 1.4 angstrom resolution crystal structure of putative alpha amylase2 from salmonella typhimurium.
|
|
|
| 86 | c3si9B_ |
|
not modelled |
47.4 |
18 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from bartonella2 henselae
|
|
|
| 87 | c1ehaA_ |
|
not modelled |
47.4 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:glycosyltrehalose trehalohydrolase;
PDBTitle: crystal structure of glycosyltrehalose trehalohydrolase from2 sulfolobus solfataricus
|
|
|
| 88 | c6mqhA_ |
|
not modelled |
47.0 |
14 |
PDB header:lyase
Chain: A: PDB Molecule:4-hydroxy-tetrahydrodipicolinate synthase;
PDBTitle: crystal structure of 4-hydroxy-tetrahydrodipicolinate synthase (htpa2 synthase) from burkholderia mallei
|
|
|
| 89 | c3px5A_ |
|
not modelled |
47.0 |
9 |
PDB header:metal binding protein
Chain: A: PDB Molecule:enzyme of enolase superfamily;
PDBTitle: structure of efi enolase target en500555, a putative dipeptide2 epimerase: apo structure
|
|
|
| 90 | c3b8iF_ |
|
not modelled |
46.7 |
29 |
PDB header:lyase
Chain: F: PDB Molecule:pa4872 oxaloacetate decarboxylase;
PDBTitle: crystal structure of oxaloacetate decarboxylase from pseudomonas2 aeruginosa (pa4872) in complex with oxalate and mg2+.
|
|
|
| 91 | c4nq1B_ |
|
not modelled |
46.2 |
11 |
PDB header:lyase
Chain: B: PDB Molecule:4-hydroxy-tetrahydrodipicolinate synthase;
PDBTitle: legionella pneumophila dihydrodipicolinate synthase with first2 substrate pyruvate bound in the active site
|
|
|
| 92 | c2p88E_ |
|
not modelled |
45.8 |
13 |
PDB header:lyase
Chain: E: PDB Molecule:mandelate racemase/muconate lactonizing enzyme
PDBTitle: crystal structure of n-succinyl arg/lys racemase from2 bacillus cereus atcc 14579
|
|
|
| 93 | c2ze3A_ |
|
not modelled |
45.5 |
24 |
PDB header:isomerase
Chain: A: PDB Molecule:dfa0005;
PDBTitle: crystal structure of dfa0005 complexed with alpha-ketoglutarate: a2 novel member of the icl/pepm superfamily from alkali-tolerant3 deinococcus ficus
|
|
|
| 94 | c2ya0A_ |
|
not modelled |
45.1 |
23 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative alkaline amylopullulanase;
PDBTitle: catalytic module of the multi-modular glycogen-degrading pneumococcal2 virulence factor spua
|
|
|
| 95 | d1uoka2 |
|
not modelled |
45.1 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
|
|
| 96 | d1ht6a2 |
|
not modelled |
44.7 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
|
|
| 97 | c2by0A_ |
|
not modelled |
44.4 |
27 |
PDB header:hydrolase
Chain: A: PDB Molecule:maltooligosyltrehalose trehalohydrolase;
PDBTitle: is radiation damage dependent on the dose-rate used during2 macromolecular crystallography data collection
|
|
|
| 98 | d1ujqa_ |
|
not modelled |
43.9 |
22 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Phosphoenolpyruvate mutase/Isocitrate lyase-like |
|
|
| 99 | c5m99A_ |
|
not modelled |
43.8 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:alpha-amylase;
PDBTitle: functional characterization and crystal structure of thermostable2 amylase from thermotoga petrophila, reveals high thermostability and3 an archaic form of dimerization
|
|
|
| 100 | d2guya2 |
|
not modelled |
43.6 |
23 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
|
|
| 101 | c2bdqA_ |
|
not modelled |
43.3 |
16 |
PDB header:metal transport
Chain: A: PDB Molecule:copper homeostasis protein cutc;
PDBTitle: crystal structure of the putative copper homeostasis protein cutc from2 streptococcus agalactiae, northeast strucural genomics target sar15.
|
|
|
| 102 | d1jdfa1 |
|
not modelled |
43.2 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Enolase C-terminal domain-like
Family:D-glucarate dehydratase-like |
|
|
| 103 | c3q45E_ |
|
not modelled |
42.9 |
14 |
PDB header:isomerase
Chain: E: PDB Molecule:mandelate racemase/muconate lactonizing enzyme family;
PDBTitle: crystal structure of dipeptide epimerase from cytophaga hutchinsonii2 complexed with mg and dipeptide d-ala-l-val
|
|
|
| 104 | d1gvia3 |
|
not modelled |
42.7 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
|
|
| 105 | c5zxgB_ |
|
not modelled |
42.6 |
19 |
PDB header:hydrolase
Chain: B: PDB Molecule:cyclic maltosyl-maltose hydrolase;
PDBTitle: cyclic alpha-maltosyl-(1-->6)-maltose hydrolase from arthrobacter2 globiformis, ligand-free form
|
|
|
| 106 | d1wufa1 |
|
not modelled |
42.5 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Enolase C-terminal domain-like
Family:D-glucarate dehydratase-like |
|
|
| 107 | c3pueA_ |
|
not modelled |
42.4 |
14 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of the complex of dhydrodipicolinate synthase from2 acinetobacter baumannii with lysine at 2.6a resolution
|
|
|
| 108 | c2yxgD_ |
|
not modelled |
42.1 |
16 |
PDB header:lyase
Chain: D: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihyrodipicolinate synthase (dapa)
|
|
|
| 109 | c2aaaA_ |
|
not modelled |
41.9 |
19 |
PDB header:glycosidase
Chain: A: PDB Molecule:alpha-amylase;
PDBTitle: calcium binding in alpha-amylases: an x-ray diffraction study at 2.12 angstroms resolution of two enzymes from aspergillus
|
|
|
| 110 | d2aaaa2 |
|
not modelled |
41.7 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
|
|
| 111 | d1wzla3 |
|
not modelled |
41.2 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
|
|
| 112 | c3faxA_ |
|
not modelled |
41.1 |
23 |
PDB header:hydrolase
Chain: A: PDB Molecule:reticulocyte binding protein;
PDBTitle: the crystal structure of gbs pullulanase sap in complex with2 maltotetraose
|
|
|
| 113 | d1o5ka_ |
|
not modelled |
41.1 |
11 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
|
|
| 114 | c4icnB_ |
|
not modelled |
41.1 |
9 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: dihydrodipicolinate synthase from shewanella benthica
|
|
|
| 115 | d1qhoa4 |
|
not modelled |
40.6 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
|
|
| 116 | c3eezA_ |
|
not modelled |
40.1 |
4 |
PDB header:isomerase
Chain: A: PDB Molecule:putative mandelate racemase/muconate lactonizing enzyme;
PDBTitle: crystal structure of a putative mandelate racemase/muconate2 lactonizing enzyme from silicibacter pomeroyi
|
|
|
| 117 | d1ea9c3 |
|
not modelled |
39.7 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
|
|
| 118 | c3zt5D_ |
|
not modelled |
39.2 |
30 |
PDB header:hydrolase
Chain: D: PDB Molecule:putative glucanohydrolase pep1a;
PDBTitle: glge isoform 1 from streptomyces coelicolor with maltose2 bound
|
|
|
| 119 | c2wcsA_ |
|
not modelled |
39.1 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:alpha amylase, catalytic region;
PDBTitle: crystal structure of debranching enzyme from nostoc2 punctiforme (npde)
|
|
|
| 120 | c4uxdC_ |
|
not modelled |
39.0 |
5 |
PDB header:lyase
Chain: C: PDB Molecule:2-dehydro-3-deoxy-d-gluconate/2-dehydro-3-deoxy-
PDBTitle: 2-keto 3-deoxygluconate aldolase from picrophilus torridus
|
|
|