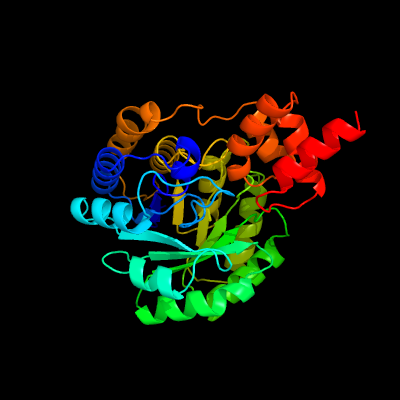
| 1 | c1nvmG_
|
|
 |
100.0 |
28 |
PDB header:lyase/oxidoreductase
Chain: G: PDB Molecule:4-hydroxy-2-oxovalerate aldolase;
PDBTitle: crystal structure of a bifunctional aldolase-dehydrogenase :2 sequestering a reactive and volatile intermediate
|
|
|
|
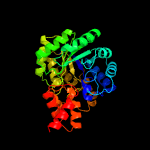
| 2 | c4jn6C_
|
|
 |
100.0 |
31 |
PDB header:lyase/oxidoreductase
Chain: C: PDB Molecule:4-hydroxy-2-oxovalerate aldolase;
PDBTitle: crystal structure of the aldolase-dehydrogenase complex from2 mycobacterium tuberculosis hrv37
|
|
|
|
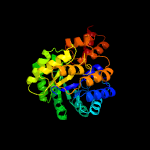
| 3 | c4lrtC_
|
|
 |
100.0 |
30 |
PDB header:lyase/oxidoreductase
Chain: C: PDB Molecule:4-hydroxy-2-oxovalerate aldolase;
PDBTitle: crystal and solution structures of the bifunctional enzyme2 (aldolase/aldehyde dehydrogenase) from thermomonospora curvata,3 reveal a cofactor-binding domain motion during nad+ and coa4 accommodation whithin the shared cofactor-binding site
|
|
|
|
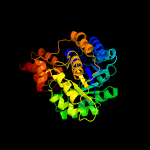
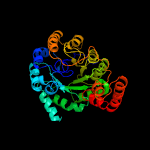
| 4 | c2nx9B_
|
|
 |
100.0 |
19 |
PDB header:lyase
Chain: B: PDB Molecule:oxaloacetate decarboxylase 2, subunit alpha;
PDBTitle: crystal structure of the carboxyltransferase domain of the2 oxaloacetate decarboxylase na+ pump from vibrio cholerae
|
|
|
|
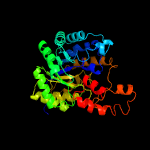
| 5 | c1rr2A_
|
|
 |
100.0 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:transcarboxylase 5s subunit;
PDBTitle: propionibacterium shermanii transcarboxylase 5s subunit bound to 2-2 ketobutyric acid
|
|
|
|
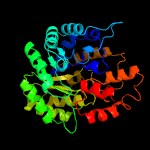
| 6 | c5ks8D_
|
|
 |
100.0 |
20 |
PDB header:ligase
Chain: D: PDB Molecule:pyruvate carboxylase subunit beta;
PDBTitle: crystal structure of two-subunit pyruvate carboxylase from2 methylobacillus flagellatus
|
|
|
|
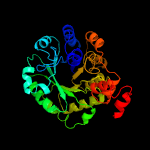
| 7 | c3dxiB_
|
|
 |
100.0 |
22 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:putative aldolase;
PDBTitle: crystal structure of the n-terminal domain of a putative2 aldolase (bvu_2661) from bacteroides vulgatus
|
|
|
|
| 8 | d1nvma2
|
|
 |
100.0 |
30 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:HMGL-like |
|
|
|
| 9 | c4qslE_
|
|
 |
100.0 |
16 |
PDB header:ligase
Chain: E: PDB Molecule:pyruvate carboxylase;
PDBTitle: crystal structure of listeria monocytogenes pyruvate carboxylase
|
|
|
|
| 10 | c3bg3A_
|
|
 |
100.0 |
17 |
PDB header:ligase
Chain: A: PDB Molecule:pyruvate carboxylase, mitochondrial;
PDBTitle: crystal structure of human pyruvate carboxylase (missing the biotin2 carboxylase domain at the n-terminus)
|
|
|
|
| 11 | c3bg5C_
|
|
 |
100.0 |
17 |
PDB header:ligase
Chain: C: PDB Molecule:pyruvate carboxylase;
PDBTitle: crystal structure of staphylococcus aureus pyruvate carboxylase
|
|
|
|
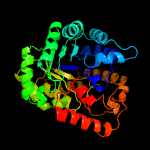
| 12 | c3bg3B_
|
|
 |
100.0 |
18 |
PDB header:ligase
Chain: B: PDB Molecule:pyruvate carboxylase, mitochondrial;
PDBTitle: crystal structure of human pyruvate carboxylase (missing the biotin2 carboxylase domain at the n-terminus)
|
|
|
|
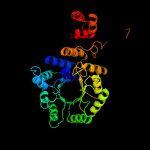
| 13 | c3ivuB_
|
|
 |
100.0 |
22 |
PDB header:transferase
Chain: B: PDB Molecule:homocitrate synthase, mitochondrial;
PDBTitle: homocitrate synthase lys4 bound to 2-og
|
|
|
|
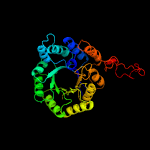
| 14 | c6e1jB_
|
|
 |
100.0 |
23 |
PDB header:plant protein
Chain: B: PDB Molecule:2-isopropylmalate synthase, a genome specific 1;
PDBTitle: crystal structure of methylthioalkylmalate synthase (bjumam1.1) from2 brassica juncea
|
|
|
|
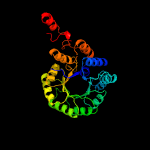
| 15 | c1sr9A_
|
|
 |
100.0 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:2-isopropylmalate synthase;
PDBTitle: crystal structure of leua from mycobacterium tuberculosis
|
|
|
|
| 16 | c3rmjB_
|
|
 |
100.0 |
24 |
PDB header:transferase
Chain: B: PDB Molecule:2-isopropylmalate synthase;
PDBTitle: crystal structure of truncated alpha-isopropylmalate synthase from2 neisseria meningitidis
|
|
|
|
| 17 | c2ftpA_
|
|
 |
100.0 |
22 |
PDB header:lyase
Chain: A: PDB Molecule:hydroxymethylglutaryl-coa lyase;
PDBTitle: crystal structure of hydroxymethylglutaryl-coa lyase from pseudomonas2 aeruginosa
|
|
|
|
| 18 | c4ov9A_
|
|
 |
100.0 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:isopropylmalate synthase;
PDBTitle: structure of isopropylmalate synthase binding with alpha-2 isopropylmalate
|
|
|
|
| 19 | c3a9iA_
|
|
 |
100.0 |
21 |
PDB header:transferase/transferase inhibitor
Chain: A: PDB Molecule:homocitrate synthase;
PDBTitle: crystal structure of homocitrate synthase from thermus thermophilus2 complexed with lys
|
|
|
|
| 20 | c3bleA_
|
|
 |
100.0 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:citramalate synthase from leptospira interrogans;
PDBTitle: crystal structure of the catalytic domain of licms in complexed with2 malonate
|
|
|
|
| 21 | c4qslC_ |
|
not modelled |
100.0 |
16 |
PDB header:ligase
Chain: C: PDB Molecule:pyruvate carboxylase;
PDBTitle: crystal structure of listeria monocytogenes pyruvate carboxylase
|
|
|
| 22 | c2zyfA_ |
|
not modelled |
100.0 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:homocitrate synthase;
PDBTitle: crystal structure of homocitrate synthase from thermus thermophilus2 complexed with magnesuim ion and alpha-ketoglutarate
|
|
|
| 23 | c1ydnA_ |
|
not modelled |
100.0 |
20 |
PDB header:lyase
Chain: A: PDB Molecule:hydroxymethylglutaryl-coa lyase;
PDBTitle: crystal structure of the hmg-coa lyase from brucella melitensis,2 northeast structural genomics target lr35.
|
|
|
| 24 | c5ks8F_ |
|
not modelled |
100.0 |
21 |
PDB header:ligase
Chain: F: PDB Molecule:pyruvate carboxylase subunit beta;
PDBTitle: crystal structure of two-subunit pyruvate carboxylase from2 methylobacillus flagellatus
|
|
|
| 25 | c2cw6B_ |
|
not modelled |
100.0 |
22 |
PDB header:lyase
Chain: B: PDB Molecule:hydroxymethylglutaryl-coa lyase, mitochondrial;
PDBTitle: crystal structure of human hmg-coa lyase: insights into2 catalysis and the molecular basis for3 hydroxymethylglutaric aciduria
|
|
|
| 26 | d1rqba2 |
|
not modelled |
100.0 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:HMGL-like |
|
|
| 27 | c3hpxB_ |
|
not modelled |
100.0 |
19 |
PDB header:transferase
Chain: B: PDB Molecule:2-isopropylmalate synthase;
PDBTitle: crystal structure of mycobacterium tuberculosis leua active site2 domain 1-425 (truncation mutant delta:426-644)
|
|
|
| 28 | c1ydoC_ |
|
not modelled |
100.0 |
20 |
PDB header:lyase
Chain: C: PDB Molecule:hmg-coa lyase;
PDBTitle: crystal structure of the bacillis subtilis hmg-coa lyase, northeast2 structural genomics target sr181.
|
|
|
| 29 | c3ewbX_ |
|
not modelled |
100.0 |
24 |
PDB header:transferase
Chain: X: PDB Molecule:2-isopropylmalate synthase;
PDBTitle: crystal structure of n-terminal domain of putative 2-isopropylmalate2 synthase from listeria monocytogenes
|
|
|
| 30 | d1sr9a2 |
|
not modelled |
100.0 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:HMGL-like |
|
|
| 31 | c3eegB_ |
|
not modelled |
100.0 |
25 |
PDB header:transferase
Chain: B: PDB Molecule:2-isopropylmalate synthase;
PDBTitle: crystal structure of a 2-isopropylmalate synthase from2 cytophaga hutchinsonii
|
|
|
| 32 | c2qf7A_ |
|
not modelled |
100.0 |
19 |
PDB header:ligase
Chain: A: PDB Molecule:pyruvate carboxylase protein;
PDBTitle: crystal structure of a complete multifunctional pyruvate carboxylase2 from rhizobium etli
|
|
|
| 33 | c4qskB_ |
|
not modelled |
100.0 |
15 |
PDB header:ligase
Chain: B: PDB Molecule:pyruvate carboxylase;
PDBTitle: crystal structure of l. monocytogenes pyruvate carboxylase in complex2 with cyclic-di-amp
|
|
|
| 34 | c4hnvB_ |
|
not modelled |
100.0 |
16 |
PDB header:ligase
Chain: B: PDB Molecule:pyruvate carboxylase;
PDBTitle: crystal structure of r54e mutant of s. aureus pyruvate carboxylase
|
|
|
| 35 | c5vz0D_ |
|
not modelled |
100.0 |
16 |
PDB header:ligase
Chain: D: PDB Molecule:pyruvate carboxylase;
PDBTitle: crystal structure of lactococcus lactis pyruvate carboxylase g746a2 mutant in complex with cyclic-di-amp
|
|
|
| 36 | c3bg5B_ |
|
not modelled |
100.0 |
17 |
PDB header:ligase
Chain: B: PDB Molecule:pyruvate carboxylase;
PDBTitle: crystal structure of staphylococcus aureus pyruvate carboxylase
|
|
|
| 37 | c3tw6B_ |
|
not modelled |
100.0 |
19 |
PDB header:ligase/activator
Chain: B: PDB Molecule:pyruvate carboxylase protein;
PDBTitle: structure of rhizobium etli pyruvate carboxylase t882a with the2 allosteric activator, acetyl coenzyme-a
|
|
|
| 38 | c3hblA_ |
|
not modelled |
100.0 |
16 |
PDB header:ligase
Chain: A: PDB Molecule:pyruvate carboxylase;
PDBTitle: crystal structure of s. aureus pyruvate carboxylase t908a mutant
|
|
|
| 39 | c2ekcA_ |
|
not modelled |
98.6 |
15 |
PDB header:lyase
Chain: A: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: structural study of project id aq_1548 from aquifex aeolicus vf5
|
|
|
| 40 | c3vndD_ |
|
not modelled |
98.5 |
17 |
PDB header:lyase
Chain: D: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: crystal structure of tryptophan synthase alpha-subunit from the2 psychrophile shewanella frigidimarina k14-2
|
|
|
| 41 | d1xcfa_ |
|
not modelled |
98.5 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
|
|
| 42 | c5kzmA_ |
|
not modelled |
98.5 |
16 |
PDB header:lyase
Chain: A: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: crystal structure of tryptophan synthase alpha-beta chain complex from2 francisella tularensis
|
|
|
| 43 | c3thaB_ |
|
not modelled |
98.5 |
14 |
PDB header:lyase
Chain: B: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: tryptophan synthase subunit alpha from campylobacter jejuni.
|
|
|
| 44 | c3navB_ |
|
not modelled |
98.4 |
17 |
PDB header:lyase
Chain: B: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: crystal structure of an alpha subunit of tryptophan synthase from2 vibrio cholerae o1 biovar el tor str. n16961
|
|
|
| 45 | d1qopa_ |
|
not modelled |
98.4 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
|
|
| 46 | c5n2pA_ |
|
not modelled |
98.2 |
14 |
PDB header:lyase
Chain: A: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: sulfolobus solfataricus tryptophan synthase a
|
|
|
| 47 | c5k9xA_ |
|
not modelled |
98.2 |
14 |
PDB header:lyase
Chain: A: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: crystal structure of tryptophan synthase alpha chain from legionella2 pneumophila subsp. pneumophila
|
|
|
| 48 | c3t7vA_ |
|
not modelled |
98.2 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:methylornithine synthase pylb;
PDBTitle: crystal structure of methylornithine synthase (pylb)
|
|
|
| 49 | d1rd5a_ |
|
not modelled |
98.2 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
|
|
| 50 | c5tchG_ |
|
not modelled |
98.2 |
18 |
PDB header:lyase
Chain: G: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: crystal structure of tryptophan synthase from m. tuberculosis -2 ligand-free form, trpa-g66v mutant
|
|
|
| 51 | c5kinC_ |
|
not modelled |
98.1 |
17 |
PDB header:lyase
Chain: C: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: crystal structure of tryptophan synthase alpha beta complex from2 streptococcus pneumoniae
|
|
|
| 52 | c4wcxC_ |
|
not modelled |
97.9 |
13 |
PDB header:lyase
Chain: C: PDB Molecule:biotin and thiamin synthesis associated;
PDBTitle: crystal structure of hydg: a maturase of the [fefe]-hydrogenase
|
|
|
| 53 | d1geqa_ |
|
not modelled |
97.9 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
|
|
| 54 | c5ey5A_ |
|
not modelled |
97.9 |
19 |
PDB header:lyase
Chain: A: PDB Molecule:lbcats-a;
PDBTitle: lbcats
|
|
|
| 55 | c3cixA_ |
|
not modelled |
97.9 |
11 |
PDB header:adomet binding protein
Chain: A: PDB Molecule:fefe-hydrogenase maturase;
PDBTitle: x-ray structure of the [fefe]-hydrogenase maturase hyde from2 thermotoga maritima in complex with thiocyanate
|
|
|
| 56 | d1ujpa_ |
|
not modelled |
97.7 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
|
|
| 57 | c5exkG_ |
|
not modelled |
97.7 |
14 |
PDB header:transferase
Chain: G: PDB Molecule:lipoyl synthase;
PDBTitle: crystal structure of m. tuberculosis lipoyl synthase with 6-2 thiooctanoyl peptide intermediate
|
|
|
| 58 | d1rvga_ |
|
not modelled |
97.6 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class II FBP aldolase |
|
|
| 59 | c5u4nA_ |
|
not modelled |
97.5 |
16 |
PDB header:lyase
Chain: A: PDB Molecule:fructose-1;
PDBTitle: crystal structure of a fructose-bisphosphate aldolase from neisseria2 gonorrhoeae
|
|
|
| 60 | c4nu7C_ |
|
not modelled |
97.5 |
13 |
PDB header:isomerase
Chain: C: PDB Molecule:ribulose-phosphate 3-epimerase;
PDBTitle: 2.05 angstrom crystal structure of ribulose-phosphate 3-epimerase from2 toxoplasma gondii.
|
|
|
| 61 | c1r30A_ |
|
not modelled |
97.5 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:biotin synthase;
PDBTitle: the crystal structure of biotin synthase, an s-adenosylmethionine-2 dependent radical enzyme
|
|
|
| 62 | d1r30a_ |
|
not modelled |
97.5 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Radical SAM enzymes
Family:Biotin synthase |
|
|
| 63 | d1nvma1 |
|
not modelled |
97.5 |
18 |
Fold:RuvA C-terminal domain-like
Superfamily:post-HMGL domain-like
Family:DmpG/LeuA communication domain-like |
|
|
| 64 | d1tqxa_ |
|
not modelled |
97.4 |
8 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:D-ribulose-5-phosphate 3-epimerase |
|
|
| 65 | c3ajxA_ |
|
not modelled |
97.4 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:3-hexulose-6-phosphate synthase;
PDBTitle: crystal structure of 3-hexulose-6-phosphate synthase
|
|
|
| 66 | c3f4wA_ |
|
not modelled |
97.4 |
13 |
PDB header:synthase, lyase
Chain: A: PDB Molecule:putative hexulose 6 phosphate synthase;
PDBTitle: the 1.65a crystal structure of 3-hexulose-6-phosphate2 synthase from salmonella typhimurium
|
|
|
| 67 | c3c52B_ |
|
not modelled |
97.3 |
12 |
PDB header:lyase
Chain: B: PDB Molecule:fructose-bisphosphate aldolase;
PDBTitle: class ii fructose-1,6-bisphosphate aldolase from helicobacter pylori2 in complex with phosphoglycolohydroxamic acid, a competitive3 inhibitor
|
|
|
| 68 | c6ncsB_ |
|
not modelled |
97.3 |
13 |
PDB header:biosynthetic protein
Chain: B: PDB Molecule:n-acetylneuraminic acid (sialic acid) synthetase;
PDBTitle: crystal structure of n-acetylneuraminic acid (sialic acid) synthetase2 from leptospira borgpetersenii serovar hardjo-bovis in complex with3 citrate
|
|
|
| 69 | c3nvtA_ |
|
not modelled |
97.3 |
15 |
PDB header:transferase/isomerase
Chain: A: PDB Molecule:3-deoxy-d-arabino-heptulosonate 7-phosphate synthase;
PDBTitle: 1.95 angstrom crystal structure of a bifunctional 3-deoxy-7-2 phosphoheptulonate synthase/chorismate mutase (aroa) from listeria3 monocytogenes egd-e
|
|
|
| 70 | c5b69A_ |
|
not modelled |
96.9 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:geranylgeranylglyceryl phosphate synthase;
PDBTitle: crystal structure of geranylgeranylglyceryl phosphate synthase2 complexed with an g-1-p from thermoplasma acidophilum
|
|
|
| 71 | c4e38A_ |
|
not modelled |
96.9 |
18 |
PDB header:lyase
Chain: A: PDB Molecule:keto-hydroxyglutarate-aldolase/keto-deoxy-phosphogluconate
PDBTitle: crystal structure of probable keto-hydroxyglutarate-aldolase from2 vibrionales bacterium swat-3 (target efi-502156)
|
|
|
| 72 | d1q6oa_ |
|
not modelled |
96.8 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Decarboxylase |
|
|
| 73 | c3jr2D_ |
|
not modelled |
96.8 |
14 |
PDB header:biosynthetic protein
Chain: D: PDB Molecule:hexulose-6-phosphate synthase sgbh;
PDBTitle: x-ray crystal structure of the mg-bound 3-keto-l-gulonate-6-phosphate2 decarboxylase from vibrio cholerae o1 biovar el tor str. n16961
|
|
|
| 74 | c2iswB_ |
|
not modelled |
96.7 |
14 |
PDB header:lyase
Chain: B: PDB Molecule:putative fructose-1,6-bisphosphate aldolase;
PDBTitle: structure of giardia fructose-1,6-biphosphate aldolase in2 complex with phosphoglycolohydroxamate
|
|
|
| 75 | c3pg8B_ |
|
not modelled |
96.6 |
11 |
PDB header:transferase
Chain: B: PDB Molecule:phospho-2-dehydro-3-deoxyheptonate aldolase;
PDBTitle: truncated form of 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase2 from thermotoga maritima
|
|
|
| 76 | c4lu0A_ |
|
not modelled |
96.6 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:2-dehydro-3-deoxyphosphooctonate aldolase;
PDBTitle: crystal structure of 2-keto-3-deoxy-d-manno-octulosonate-8-phosphate2 synthase from pseudomonas aeruginosa.
|
|
|
| 77 | d1h1ya_ |
|
not modelled |
96.5 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:D-ribulose-5-phosphate 3-epimerase |
|
|
| 78 | c6oviA_ |
|
not modelled |
96.5 |
12 |
PDB header:lyase
Chain: A: PDB Molecule:keto-deoxy-phosphogluconate aldolase;
PDBTitle: crystal structure of kdpg aldolase from legionella pneumophila with2 pyruvate captured at low ph as a covalent carbinolamine intermediate
|
|
|
| 79 | d1vr6a1 |
|
not modelled |
96.4 |
11 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I DAHP synthetase |
|
|
| 80 | d1ka9f_ |
|
not modelled |
96.4 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Histidine biosynthesis enzymes |
|
|
| 81 | c3inpA_ |
|
not modelled |
96.4 |
14 |
PDB header:isomerase
Chain: A: PDB Molecule:d-ribulose-phosphate 3-epimerase;
PDBTitle: 2.05 angstrom resolution crystal structure of d-ribulose-phosphate 3-2 epimerase from francisella tularensis.
|
|
|
| 82 | c3qc3B_ |
|
not modelled |
96.4 |
11 |
PDB header:isomerase
Chain: B: PDB Molecule:d-ribulose-5-phosphate-3-epimerase;
PDBTitle: crystal structure of a d-ribulose-5-phosphate-3-epimerase (np_954699)2 from homo sapiens at 2.20 a resolution
|
|
|
| 83 | d1thfd_ |
|
not modelled |
96.4 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Histidine biosynthesis enzymes |
|
|
| 84 | d2zdra2 |
|
not modelled |
96.4 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:NeuB-like |
|
|
| 85 | d1gvfa_ |
|
not modelled |
96.4 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class II FBP aldolase |
|
|
| 86 | c3pm6B_ |
|
not modelled |
96.3 |
16 |
PDB header:lyase
Chain: B: PDB Molecule:putative fructose-bisphosphate aldolase;
PDBTitle: crystal structure of a putative fructose-1,6-biphosphate aldolase from2 coccidioides immitis solved by combined sad mr
|
|
|
| 87 | d1vhca_ |
|
not modelled |
96.2 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
|
|
| 88 | c1vliA_ |
|
not modelled |
96.2 |
13 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:spore coat polysaccharide biosynthesis protein spse;
PDBTitle: crystal structure of spore coat polysaccharide biosynthesis protein2 spse (bsu37870) from bacillus subtilis at 2.38 a resolution
|
|
|
| 89 | c2ou4C_ |
|
not modelled |
96.2 |
16 |
PDB header:isomerase
Chain: C: PDB Molecule:d-tagatose 3-epimerase;
PDBTitle: crystal structure of d-tagatose 3-epimerase from2 pseudomonas cichorii
|
|
|
| 90 | d1mxsa_ |
|
not modelled |
96.2 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
|
|
| 91 | c4rtbA_ |
|
not modelled |
96.2 |
14 |
PDB header:lyase
Chain: A: PDB Molecule:hydg protein;
PDBTitle: x-ray structure of the fefe-hydrogenase maturase hydg from2 carboxydothermus hydrogenoformans
|
|
|
| 92 | c6nkeA_ |
|
not modelled |
96.1 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:geranylgeranylglyceryl phosphate synthase;
PDBTitle: wild-type gggps from thermoplasma volcanium
|
|
|
| 93 | d1tqja_ |
|
not modelled |
96.0 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:D-ribulose-5-phosphate 3-epimerase |
|
|
| 94 | d1wbha1 |
|
not modelled |
95.9 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
|
|
| 95 | d1vlia2 |
|
not modelled |
95.9 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:NeuB-like |
|
|
| 96 | c4ml9A_ |
|
not modelled |
95.9 |
22 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of uncharacterized tim barrel protein with the2 conserved phosphate binding site fromsebaldella termitidis
|
|
|
| 97 | c3exsB_ |
|
not modelled |
95.8 |
14 |
PDB header:lyase
Chain: B: PDB Molecule:rmpd (hexulose-6-phosphate synthase);
PDBTitle: crystal structure of kgpdc from streptococcus mutans in2 complex with d-r5p
|
|
|
| 98 | c3vylB_ |
|
not modelled |
95.8 |
18 |
PDB header:isomerase
Chain: B: PDB Molecule:l-ribulose 3-epimerase;
PDBTitle: structure of l-ribulose 3-epimerase
|
|
|
| 99 | c1zcoA_ |
|
not modelled |
95.7 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:2-dehydro-3-deoxyphosphoheptonate aldolase;
PDBTitle: crystal structure of pyrococcus furiosus 3-deoxy-d-arabino-2 heptulosonate 7-phosphate synthase
|
|
|
| 100 | c3w9zA_ |
|
not modelled |
95.7 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:trna-dihydrouridine synthase c;
PDBTitle: crystal structure of dusc
|
|
|
| 101 | d2flia1 |
|
not modelled |
95.7 |
11 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:D-ribulose-5-phosphate 3-epimerase |
|
|
| 102 | c1tx2A_ |
|
not modelled |
95.6 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:dhps, dihydropteroate synthase;
PDBTitle: dihydropteroate synthetase, with bound inhibitor manic, from bacillus2 anthracis
|
|
|
| 103 | d1tx2a_ |
|
not modelled |
95.6 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Dihydropteroate synthetase-like
Family:Dihydropteroate synthetase |
|
|
| 104 | c3ct7E_ |
|
not modelled |
95.6 |
11 |
PDB header:isomerase
Chain: E: PDB Molecule:d-allulose-6-phosphate 3-epimerase;
PDBTitle: crystal structure of d-allulose 6-phosphate 3-epimerase2 from escherichia coli k-12
|
|
|
| 105 | c1xuzA_ |
|
not modelled |
95.5 |
16 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:polysialic acid capsule biosynthesis protein siac;
PDBTitle: crystal structure analysis of sialic acid synthase (neub)from2 neisseria meningitidis, bound to mn2+, phosphoenolpyruvate, and n-3 acetyl mannosaminitol
|
|
|
| 106 | c5d88A_ |
|
not modelled |
95.5 |
25 |
PDB header:hydrolase
Chain: A: PDB Molecule:predicted protease of the collagenase family;
PDBTitle: the structure of the u32 peptidase mk0906
|
|
|
| 107 | c3vkbA_ |
|
not modelled |
95.4 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:moeo5;
PDBTitle: crystal structure of moeo5 soaked with fspp overnight
|
|
|
| 108 | c3elfA_ |
|
not modelled |
95.3 |
15 |
PDB header:lyase
Chain: A: PDB Molecule:fructose-bisphosphate aldolase;
PDBTitle: structural characterization of tetrameric mycobacterium tuberculosis2 fructose 1,6-bisphosphate aldolase - substrate binding and catalysis3 mechanism of a class iia bacterial aldolase
|
|
|
| 109 | c3sz8D_ |
|
not modelled |
95.3 |
14 |
PDB header:transferase
Chain: D: PDB Molecule:2-dehydro-3-deoxyphosphooctonate aldolase 2;
PDBTitle: crystal structure of 2-dehydro-3-deoxyphosphooctonate aldolase from2 burkholderia pseudomallei
|
|
|
| 110 | d1d9ea_ |
|
not modelled |
95.2 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I DAHP synthetase |
|
|
| 111 | c5tnvA_ |
|
not modelled |
95.1 |
16 |
PDB header:isomerase
Chain: A: PDB Molecule:ap endonuclease, family protein 2;
PDBTitle: crystal structure of a xylose isomerase-like tim barrel protein from2 mycobacterium smegmatis in complex with magnesium
|
|
|
| 112 | c2h9aB_ |
|
not modelled |
95.1 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:co dehydrogenase/acetyl-coa synthase, iron-sulfur protein;
PDBTitle: corrinoid iron-sulfur protein
|
|
|
| 113 | c2c3zA_ |
|
not modelled |
95.0 |
15 |
PDB header:lyase
Chain: A: PDB Molecule:indole-3-glycerol phosphate synthase;
PDBTitle: crystal structure of a truncated variant of indole-3-2 glycerol phosphate synthase from sulfolobus solfataricus
|
|
|
| 114 | d1qwga_ |
|
not modelled |
95.0 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:(2r)-phospho-3-sulfolactate synthase ComA
Family:(2r)-phospho-3-sulfolactate synthase ComA |
|
|
| 115 | c1gthD_ |
|
not modelled |
94.9 |
14 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:dihydropyrimidine dehydrogenase;
PDBTitle: dihydropyrimidine dehydrogenase (dpd) from pig, ternary complex with2 nadph and 5-iodouracil
|
|
|
| 116 | d1tv8a_ |
|
not modelled |
94.9 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Radical SAM enzymes
Family:MoCo biosynthesis proteins |
|
|
| 117 | c4mwaA_ |
|
not modelled |
94.9 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase;
PDBTitle: 1.85 angstrom crystal structure of gcpe protein from bacillus2 anthracis
|
|
|
| 118 | c4u0pB_ |
|
not modelled |
94.9 |
11 |
PDB header:transferase
Chain: B: PDB Molecule:lipoyl synthase 2;
PDBTitle: the crystal structure of lipoyl synthase in complex with s-adenosyl2 homocysteine
|
|
|
| 119 | c3bolB_ |
|
not modelled |
94.8 |
14 |
PDB header:transferase
Chain: B: PDB Molecule:5-methyltetrahydrofolate s-homocysteine
PDBTitle: cobalamin-dependent methionine synthase (1-566) from2 thermotoga maritima complexed with zn2+
|
|
|
| 120 | c3l23A_ |
|
not modelled |
94.8 |
11 |
PDB header:isomerase
Chain: A: PDB Molecule:sugar phosphate isomerase/epimerase;
PDBTitle: crystal structure of sugar phosphate isomerase/epimerase2 (yp_001303399.1) from parabacteroides distasonis atcc 8503 at 1.70 a3 resolution
|
|
|