| 1 |
|
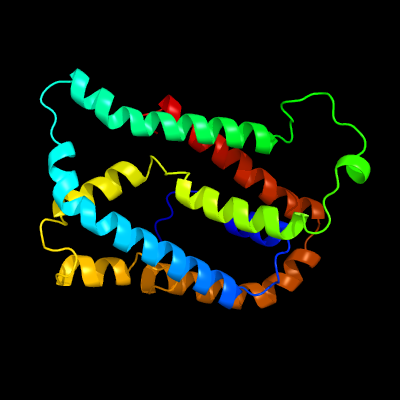
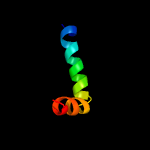
PDB 5vkv chain A
Region: 22 - 229
Aligned: 190
Modelled: 208
Confidence: 99.2%
Identity: 23%
PDB header:oxidoreductase
Chain: A: PDB Molecule:cytochrome c-type biogenesis protein ccda;
PDBTitle: solution nmr structure of the membrane electron transporter ccda
Phyre2
| 2 |
|
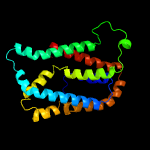
PDB 2n4x chain A
Region: 22 - 226
Aligned: 179
Modelled: 205
Confidence: 97.8%
Identity: 11%
PDB header:membrane protein
Chain: A: PDB Molecule:cytochrome c-type biogenesis protein (ccda);
PDBTitle: structure of the transmembrane electron transporter ccda
Phyre2
| 3 |
|
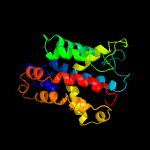
PDB 4zdt chain B
Region: 195 - 206
Aligned: 12
Modelled: 12
Confidence: 23.6%
Identity: 17%
PDB header:hydrolase
Chain: B: PDB Molecule:structure-specific endonuclease subunit slx4;
PDBTitle: crystal structure of the ring finger domain of slx1 in complex with2 the c-terminal domain of slx4
Phyre2
| 4 |
|
PDB 4ni3 chain B
Region: 188 - 209
Aligned: 22
Modelled: 22
Confidence: 14.7%
Identity: 9%
PDB header:hydrolase
Chain: B: PDB Molecule:alpha-fucosidase gh29;
PDBTitle: crystal structure of gh29 family alpha-l-fucosidase from fusarium2 graminearum in the closed form
Phyre2
| 5 |
|
PDB 5mb9 chain C
Region: 1 - 13
Aligned: 13
Modelled: 13
Confidence: 12.8%
Identity: 46%
PDB header:chaperone
Chain: C: PDB Molecule:putative ribosome associated protein;
PDBTitle: crystal structure of the eukaryotic ribosome associated complex (rac),2 a unique hsp70/hsp40 pair
Phyre2
| 6 |
|
PDB 2knc chain A
Region: 71 - 113
Aligned: 43
Modelled: 43
Confidence: 12.7%
Identity: 28%
PDB header:cell adhesion
Chain: A: PDB Molecule:integrin alpha-iib;
PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
Phyre2
| 7 |
|
PDB 1p58 chain E
Region: 187 - 216
Aligned: 30
Modelled: 30
Confidence: 12.0%
Identity: 10%
PDB header:virus
Chain: E: PDB Molecule:envelope protein m;
PDBTitle: complex organization of dengue virus membrane proteins as revealed by2 9.5 angstrom cryo-em reconstruction
Phyre2
| 8 |
|
PDB 5mb9 chain D
Region: 1 - 13
Aligned: 13
Modelled: 13
Confidence: 10.7%
Identity: 46%
PDB header:chaperone
Chain: D: PDB Molecule:putative ribosome associated protein;
PDBTitle: crystal structure of the eukaryotic ribosome associated complex (rac),2 a unique hsp70/hsp40 pair
Phyre2
| 9 |
|
PDB 1p58 chain F
Region: 187 - 214
Aligned: 28
Modelled: 28
Confidence: 10.2%
Identity: 11%
PDB header:virus
Chain: F: PDB Molecule:envelope protein m;
PDBTitle: complex organization of dengue virus membrane proteins as revealed by2 9.5 angstrom cryo-em reconstruction
Phyre2
| 10 |
|
PDB 2lkl chain A
Region: 195 - 208
Aligned: 14
Modelled: 14
Confidence: 9.1%
Identity: 21%
PDB header:cell adhesion
Chain: A: PDB Molecule:erythrocyte membrane protein 1 (pfemp1);
PDBTitle: structure of the core intracellular domain of pfemp1
Phyre2
| 11 |
|
PDB 6gn6 chain A
Region: 188 - 209
Aligned: 22
Modelled: 22
Confidence: 8.5%
Identity: 23%
PDB header:hydrolase
Chain: A: PDB Molecule:alpha-l-fucosidase;
PDBTitle: alpha-l-fucosidase isoenzyme 1 from paenibacillus thiaminolyticus
Phyre2
| 12 |
|
PDB 3bjr chain A
Region: 188 - 206
Aligned: 19
Modelled: 19
Confidence: 8.5%
Identity: 16%
PDB header:hydrolase
Chain: A: PDB Molecule:putative carboxylesterase;
PDBTitle: crystal structure of a putative carboxylesterase (lp_1002) from2 lactobacillus plantarum wcfs1 at 2.09 a resolution
Phyre2
| 13 |
|
PDB 2g5d chain A domain 1
Region: 195 - 213
Aligned: 19
Modelled: 19
Confidence: 7.0%
Identity: 16%
Fold: Double psi beta-barrel
Superfamily: Barwin-like endoglucanases
Family: MLTA-like
Phyre2
| 14 |
|
PDB 4b03 chain D
Region: 187 - 216
Aligned: 30
Modelled: 30
Confidence: 6.9%
Identity: 10%
PDB header:virus
Chain: D: PDB Molecule:dengue virus 1 prm protein;
PDBTitle: 6a electron cryomicroscopy structure of immature dengue virus serotype2 1
Phyre2
| 15 |
|
PDB 2pnw chain A domain 1
Region: 195 - 213
Aligned: 19
Modelled: 19
Confidence: 6.5%
Identity: 5%
Fold: Double psi beta-barrel
Superfamily: Barwin-like endoglucanases
Family: MLTA-like
Phyre2
| 16 |
|
PDB 2auw chain A domain 1
Region: 196 - 206
Aligned: 11
Modelled: 9
Confidence: 6.5%
Identity: 36%
Fold: lambda repressor-like DNA-binding domains
Superfamily: lambda repressor-like DNA-binding domains
Family: NE0471 C-terminal domain-like
Phyre2
| 17 |
|
PDB 4eq7 chain B
Region: 189 - 207
Aligned: 19
Modelled: 19
Confidence: 5.6%
Identity: 32%
PDB header:transport protein
Chain: B: PDB Molecule:abc transporter, substrate binding protein (polyamine);
PDBTitle: structure of atu4243-gaba receptor
Phyre2
| 18 |
|
PDB 6mjp chain F
Region: 23 - 118
Aligned: 94
Modelled: 96
Confidence: 5.4%
Identity: 10%
PDB header:lipid transport
Chain: F: PDB Molecule:fig000988: predicted permease;
PDBTitle: lptb(e163q)fgc from vibrio cholerae
Phyre2
| 19 |
|
PDB 2mqk chain A
Region: 188 - 206
Aligned: 19
Modelled: 19
Confidence: 5.4%
Identity: 26%
PDB header:hydrolase
Chain: A: PDB Molecule:atp-dependent target dna activator b;
PDBTitle: solution structure of n terminal domain of the mub aaa+ atpase
Phyre2
| 20 |
|
PDB 3j2p chain D
Region: 187 - 216
Aligned: 30
Modelled: 30
Confidence: 5.3%
Identity: 10%
PDB header:viral protein
Chain: D: PDB Molecule:small envelope protein m;
PDBTitle: cryoem structure of dengue virus envelope protein heterotetramer
Phyre2
| 21 |
|
| 22 |
|