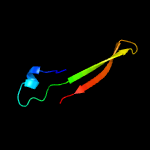
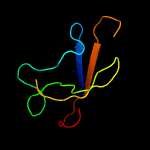
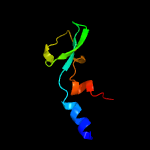
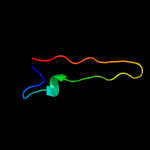
1 c4r7vA_
43.3
17
PDB header: protein bindingChain: A: PDB Molecule: arrestin domain-containing protein 3;PDBTitle: crystal structure of n-lobe of human arrdc3(1-165)
2 c6czjA_
31.8
38
PDB header: de novo proteinChain: A: PDB Molecule: b10;PDBTitle: structure of a redesigned beta barrel, b10
3 d1hxma2
27.6
26
Fold: Immunoglobulin-like beta-sandwichSuperfamily: ImmunoglobulinFamily: C1 set domains (antibody constant domain-like)
4 c3qrzC_
22.3
20
PDB header: hormone receptorChain: C: PDB Molecule: abscisic acid receptor pyl5;PDBTitle: crystal structure of native abscisic acid receptor pyl5 at 2.62 angstrom
5 c1g4mA_
19.6
19
PDB header: signaling proteinChain: A: PDB Molecule: beta-arrestin1;PDBTitle: crystal structure of bovine beta-arrestin 1
6 c5icuA_
17.6
12
PDB header: chaperoneChain: A: PDB Molecule: copc;PDBTitle: the crystal structure of copc from methylosinus trichosporium ob3b
7 c4gejD_
17.3
14
PDB header: protein bindingChain: D: PDB Molecule: thioredoxin-interacting protein;PDBTitle: n-terminal domain of vdup-1
8 c1sujA_
16.8
19
PDB header: signaling proteinChain: A: PDB Molecule: cone arrestin;PDBTitle: x-ray crystal structure of ambystoma tigrinum cone arrestin
9 c3n6yA_
16.7
23
PDB header: unknown functionChain: A: PDB Molecule: immunoglobulin-like protein;PDBTitle: crystal structure of an immunoglobulin-like protein (pa1606) from2 pseudomonas aeruginosa at 1.50 a resolution
10 c1ayrA_
15.8
25
PDB header: sensory transductionChain: A: PDB Molecule: arrestin;PDBTitle: arrestin from bovine rod outer segments
11 c1jsyA_
15.6
20
PDB header: signaling proteinChain: A: PDB Molecule: bovine arrestin-2 (full length);PDBTitle: crystal structure of bovine arrestin-2
12 c5djjA_
14.5
25
PDB header: hydrolaseChain: A: PDB Molecule: 3'-phosphoadenosine 5'-phosphate phosphatase;PDBTitle: structure of m. tuberculosis cysq, a pap phosphatase with po4 and 2mg2 bound
13 c5x5sA_
14.2
15
PDB header: hydrolaseChain: A: PDB Molecule: amylase;PDBTitle: ligand induced structure of amyp-sbd
14 d2q9oa3
12.3
21
Fold: Cupredoxin-likeSuperfamily: CupredoxinsFamily: Multidomain cupredoxins
15 d2hg6a1
12.1
24
Fold: PA1123-likeSuperfamily: PA1123-likeFamily: PA1123-like
16 c1cf1B_
11.6
25
PDB header: structural proteinChain: B: PDB Molecule: protein (arrestin);PDBTitle: arrestin from bovine rod outer segments
17 c3uc0B_
9.7
25
PDB header: viral protein/immune systemChain: B: PDB Molecule: envelope protein;PDBTitle: crystal structure of domain i of the envelope glycoprotein ectodomain2 from dengue virus serotype 4 in complex with the fab fragment of the3 chimpanzee monoclonal antibody 5h2
18 c3nrlB_
9.3
18
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein rumgna_01417;PDBTitle: crystal structure of protein rumgna_01417 from ruminococcus gnavus,2 northeast structural genomics consortium target ugr76
19 c6h7wC_
9.1
5
PDB header: protein transportChain: C: PDB Molecule: vacuolar protein sorting-associated protein 26-likePDBTitle: model of retromer-vps5 complex assembled on membrane.
20 c2rpsA_
8.9
67
PDB header: immune systemChain: A: PDB Molecule: chemokine;PDBTitle: solution structure of a novel insect chemokine isolated from2 integument
21 c6b0nG_
not modelled
8.7
27
PDB header: viral protein/immune systemChain: G: PDB Molecule: envelope glycoprotein gp140;PDBTitle: crystal structure of the cleavage-independent prefusion hiv env2 glycoprotein trimer of the clade a bg505 isolate (nfl construct) in3 complex with fabs pgt122 and pgv19 at 3.39 a
22 d1ypze2
not modelled
8.7
32
Fold: Immunoglobulin-like beta-sandwichSuperfamily: ImmunoglobulinFamily: C1 set domains (antibody constant domain-like)
23 c3fk4A_
not modelled
8.7
14
PDB header: isomeraseChain: A: PDB Molecule: rubisco-like protein;PDBTitle: crystal structure of rubisco-like protein from bacillus2 cereus atcc 14579
24 d1lmia_
not modelled
8.3
13
Fold: Immunoglobulin-like beta-sandwichSuperfamily: Antigen MPT63/MPB63 (immunoprotective extracellular protein)Family: Antigen MPT63/MPB63 (immunoprotective extracellular protein)
25 d2j5la1
not modelled
8.2
28
Fold: Apical membrane antigen 1Superfamily: Apical membrane antigen 1Family: Apical membrane antigen 1
26 c2j5lA_
not modelled
8.2
28
PDB header: immune systemChain: A: PDB Molecule: apical membrane antigen 1;PDBTitle: structure of a plasmodium falciparum apical membrane antigen 1-fab f8.2 12.19 complex
27 d1usha1
not modelled
8.2
11
Fold: 5'-nucleotidase (syn. UDP-sugar hydrolase), C-terminal domainSuperfamily: 5'-nucleotidase (syn. UDP-sugar hydrolase), C-terminal domainFamily: 5'-nucleotidase (syn. UDP-sugar hydrolase), C-terminal domain
28 c5hycC_
not modelled
8.0
47
PDB header: unknown functionChain: C: PDB Molecule: cytoplasmic dynein 1 intermediate chain 2;PDBTitle: structure based function annotation of a hypothetical protein2 mgg_01005 related to the development of rice blast fungus
29 d1ykwa1
not modelled
7.9
10
Fold: TIM beta/alpha-barrelSuperfamily: RuBisCo, C-terminal domainFamily: RuBisCo, large subunit, C-terminal domain
30 d1fcda2
not modelled
7.7
18
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: FAD/NAD-linked reductases, N-terminal and central domains
31 d1lgbc_
not modelled
7.5
31
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Transferrin
32 d1qasa3
not modelled
7.4
27
Fold: TIM beta/alpha-barrelSuperfamily: PLC-like phosphodiesterasesFamily: Mammalian PLC
33 c6cp8B_
not modelled
7.4
50
PDB header: toxin/antitoxinChain: B: PDB Molecule: cdia;PDBTitle: contact-dependent growth inhibition toxin-immunity protein complex2 from from e. coli 3006
34 d2cpqa1
not modelled
7.4
5
Fold: Eukaryotic type KH-domain (KH-domain type I)Superfamily: Eukaryotic type KH-domain (KH-domain type I)Family: Eukaryotic type KH-domain (KH-domain type I)
35 c4ufqB_
not modelled
7.1
26
PDB header: hydrolaseChain: B: PDB Molecule: hyaluronidase;PDBTitle: structure of a novel hyaluronidase (hyal_sk) from streptomyces2 koganeiensis.
36 d1ka1a_
not modelled
6.9
28
Fold: Carbohydrate phosphataseSuperfamily: Carbohydrate phosphataseFamily: Inositol monophosphatase/fructose-1,6-bisphosphatase-like
37 c5j6yA_
not modelled
6.9
22
PDB header: cell adhesionChain: A: PDB Molecule: antifreeze protein;PDBTitle: crystal structure of pa14 domain of mpafp antifreeze protein
38 c5fk0F_
not modelled
6.5
9
PDB header: structural proteinChain: F: PDB Molecule: coatomer subunit delta;PDBTitle: yeast delta-cop-i mu-homology domain
39 d1svda1
not modelled
6.4
24
Fold: TIM beta/alpha-barrelSuperfamily: RuBisCo, C-terminal domainFamily: RuBisCo, large subunit, C-terminal domain
40 c2djyB_
not modelled
6.3
46
PDB header: ligase/signaling proteinChain: B: PDB Molecule: mothers against decapentaplegic homolog 7;PDBTitle: solution structure of smurf2 ww3 domain-smad7 py peptide2 complex
41 c2kxqB_
not modelled
6.3
46
PDB header: protein bindingChain: B: PDB Molecule: smad7 py motif containing peptide;PDBTitle: solution structure of smurf2 ww2 and ww3 bound to smad7 py motif2 containing peptide
42 c5yt6B_
not modelled
6.0
17
PDB header: protein bindingChain: B: PDB Molecule: tax1-binding protein 1;PDBTitle: crystal structure of tax1bp1 ubz2 in complex with mono-ubiquitin
43 d1x4ka1
not modelled
6.0
56
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: LIM domain
44 c2czhB_
not modelled
5.9
25
PDB header: hydrolaseChain: B: PDB Molecule: inositol monophosphatase 2;PDBTitle: crystal structure of human myo-inositol monophosphatase 22 (impa2) with phosphate ion (orthorhombic form)
45 c2n59A_
not modelled
5.9
16
PDB header: unknown functionChain: A: PDB Molecule: putative uncharacterized protein csgh;PDBTitle: solution structure of r. palustris csgh
46 c6hunA_
not modelled
5.8
13
PDB header: photosynthesisChain: A: PDB Molecule: ribulose bisphosphate carboxylase;PDBTitle: dimeric archeal rubisco from hyperthermus butylicus
47 d1ej7l1
not modelled
5.8
21
Fold: TIM beta/alpha-barrelSuperfamily: RuBisCo, C-terminal domainFamily: RuBisCo, large subunit, C-terminal domain
48 d1gv8a_
not modelled
5.7
19
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Transferrin
49 c4nasD_
not modelled
5.7
21
PDB header: lyaseChain: D: PDB Molecule: ribulose-bisphosphate carboxylase;PDBTitle: the crystal structure of a rubisco-like protein (mtnw) from2 alicyclobacillus acidocaldarius subsp. acidocaldarius dsm 446
50 d1cc3a_
not modelled
5.5
13
Fold: Cupredoxin-likeSuperfamily: CupredoxinsFamily: Plastocyanin/azurin-like
51 d2fmra_
not modelled
5.5
6
Fold: Eukaryotic type KH-domain (KH-domain type I)Superfamily: Eukaryotic type KH-domain (KH-domain type I)Family: Eukaryotic type KH-domain (KH-domain type I)
52 d1w7ca2
not modelled
5.5
38
Fold: Cystatin-likeSuperfamily: Amine oxidase N-terminal regionFamily: Amine oxidase N-terminal region
53 c5swkC_
not modelled
5.4
13
PDB header: de novo proteinChain: C: PDB Molecule: de novo protein based on the inhibitor amoebiasin-1;PDBTitle: crystal structure of p53 epitope-scaffold based on a inhibitor of2 cysteine proteases in complex with human mdm2
54 c1m1jA_
not modelled
5.3
38
PDB header: blood clottingChain: A: PDB Molecule: fibrinogen alpha subunit;PDBTitle: crystal structure of native chicken fibrinogen with two different2 bound ligands
55 c4gdgA_
not modelled
5.2
21
PDB header: hydrolaseChain: A: PDB Molecule: 3'(2'),5'-bisphosphate nucleotidase, putative;PDBTitle: structural and functional characterization of 3'(2'),5'-bisphosphate2 nucleotidase1 from entamoeba histolytica
56 c3nwrA_
not modelled
5.2
17
PDB header: lyaseChain: A: PDB Molecule: a rubisco-like protein;PDBTitle: crystal structure of a rubisco-like protein from burkholderia fungorum
57 d1nwpa_
not modelled
5.2
13
Fold: Cupredoxin-likeSuperfamily: CupredoxinsFamily: Plastocyanin/azurin-like
58 c5muyB_
not modelled
5.1
38
PDB header: viral proteinChain: B: PDB Molecule: l protein;PDBTitle: structure of a c-terminal domain of a reptarenavirus l protein with2 m7gtp
59 c4h69A_
not modelled
5.1
33
PDB header: isomeraseChain: A: PDB Molecule: allene oxide cyclase;PDBTitle: crystal structure of the allene oxide cyclase 2 from physcomitrella2 patens complexed with substrate analog
60 c4s3lA_
not modelled
5.1
13
PDB header: cell adhesionChain: A: PDB Molecule: major pilin protein;PDBTitle: crystal structure of major pilin protein pitb from type ii pilus of2 streptococcus pneumoniae
61 c5lc5A_
not modelled
5.1
38
PDB header: oxidoreductaseChain: A: PDB Molecule: nadh-ubiquinone oxidoreductase chain 3;PDBTitle: structure of mammalian respiratory complex i, class2