| 1 |
|
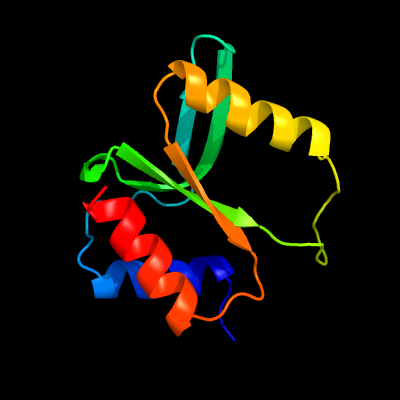
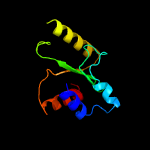
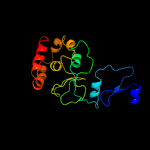
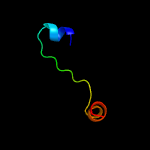
PDB 3r3p chain B
Region: 177 - 277
Aligned: 93
Modelled: 101
Confidence: 99.7%
Identity: 15%
PDB header:hydrolase
Chain: B: PDB Molecule:mobile intron protein;
PDBTitle: homing endonuclease i-bth0305i catalytic domain
Phyre2
| 2 |
|
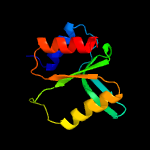
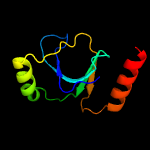
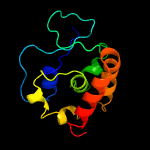
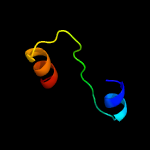
PDB 3hrl chain A
Region: 176 - 278
Aligned: 89
Modelled: 103
Confidence: 99.4%
Identity: 19%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:endonuclease-like protein;
PDBTitle: crystal structure of a putative endonuclease-like protein (ngo0050)2 from neisseria gonorrhoeae
Phyre2
| 3 |
|
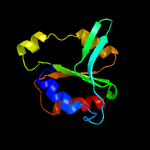
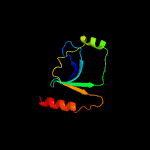
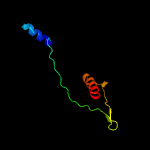
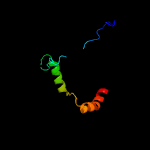
PDB 1cw0 chain A
Region: 175 - 279
Aligned: 98
Modelled: 105
Confidence: 99.3%
Identity: 18%
Fold: Restriction endonuclease-like
Superfamily: Restriction endonuclease-like
Family: Very short patch repair (VSR) endonuclease
Phyre2
| 4 |
|
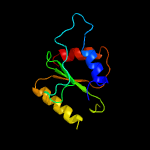
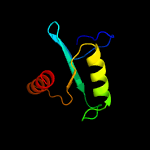
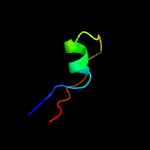
PDB 1vsr chain A
Region: 181 - 279
Aligned: 92
Modelled: 99
Confidence: 98.1%
Identity: 18%
Fold: Restriction endonuclease-like
Superfamily: Restriction endonuclease-like
Family: Very short patch repair (VSR) endonuclease
Phyre2
| 5 |
|
PDB 4oq2 chain A
Region: 195 - 278
Aligned: 84
Modelled: 84
Confidence: 96.5%
Identity: 12%
PDB header:hydrolase
Chain: A: PDB Molecule:restriction endonuclease pvurts1 i;
PDBTitle: 5hmc specific restriction endonuclease pvurts1i
Phyre2
| 6 |
|
PDB 4par chain C
Region: 195 - 278
Aligned: 82
Modelled: 84
Confidence: 96.1%
Identity: 22%
PDB header:dna binding protein/dna
Chain: C: PDB Molecule:uncharacterized protein abasi;
PDBTitle: the 5-hydroxymethylcytosine-specific restriction enzyme abasi in a2 complex with product-like dna
Phyre2
| 7 |
|
PDB 6rdu chain 2
Region: 204 - 277
Aligned: 74
Modelled: 74
Confidence: 95.7%
Identity: 12%
PDB header:proton transport
Chain: 2: PDB Molecule:asa-2: polytomella f-atp synthase associated subunit 2;
PDBTitle: cryo-em structure of polytomella f-atp synthase, rotary substate 1e,2 monomer-masked refinement
Phyre2
| 8 |
|
PDB 1zel chain A
Region: 7 - 177
Aligned: 162
Modelled: 171
Confidence: 81.7%
Identity: 20%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein rv2827c;
PDBTitle: crystal structure of rv2827c protein from mycobacterium tuberculosis
Phyre2
| 9 |
|
PDB 1zel chain A domain 2
Region: 55 - 177
Aligned: 117
Modelled: 123
Confidence: 74.9%
Identity: 19%
Fold: Rv2827c C-terminal domain-like
Superfamily: Rv2827c C-terminal domain-like
Family: Rv2827c C-terminal domain-like
Phyre2
| 10 |
|
PDB 1m0d chain A
Region: 178 - 250
Aligned: 65
Modelled: 73
Confidence: 59.9%
Identity: 17%
Fold: Restriction endonuclease-like
Superfamily: Restriction endonuclease-like
Family: Endonuclease I (Holliday junction resolvase)
Phyre2
| 11 |
|
PDB 2fcl chain A domain 1
Region: 56 - 81
Aligned: 26
Modelled: 26
Confidence: 25.0%
Identity: 15%
Fold: Nucleotidyltransferase
Superfamily: Nucleotidyltransferase
Family: TM1012-like
Phyre2
| 12 |
|
PDB 4ytk chain A
Region: 23 - 33
Aligned: 11
Modelled: 11
Confidence: 11.6%
Identity: 27%
PDB header:transcription
Chain: A: PDB Molecule:transcription elongation factor spt5;
PDBTitle: structure of the kow1-linker1 domain of transcription elongation2 factor spt5
Phyre2
| 13 |
|
PDB 2fuk chain A domain 1
Region: 192 - 258
Aligned: 67
Modelled: 67
Confidence: 11.2%
Identity: 21%
Fold: alpha/beta-Hydrolases
Superfamily: alpha/beta-Hydrolases
Family: Atu1826-like
Phyre2
| 14 |
|
PDB 1tfe chain A
Region: 219 - 277
Aligned: 59
Modelled: 59
Confidence: 10.7%
Identity: 10%
Fold: EF-Ts domain-like
Superfamily: Elongation factor Ts (EF-Ts), dimerisation domain
Family: Elongation factor Ts (EF-Ts), dimerisation domain
Phyre2
| 15 |
|
PDB 2hw2 chain A
Region: 220 - 256
Aligned: 36
Modelled: 37
Confidence: 10.6%
Identity: 8%
PDB header:transferase
Chain: A: PDB Molecule:rifampin adp-ribosyl transferase;
PDBTitle: crystal structure of rifampin adp-ribosyl transferase in complex with2 rifampin
Phyre2
| 16 |
|
PDB 4gqz chain B
Region: 193 - 229
Aligned: 37
Modelled: 36
Confidence: 9.5%
Identity: 22%
PDB header:metal binding protein
Chain: B: PDB Molecule:putative periplasmic or exported protein;
PDBTitle: crystal structure of s.cuep
Phyre2
| 17 |
|
PDB 1jg5 chain A
Region: 246 - 259
Aligned: 14
Modelled: 14
Confidence: 9.0%
Identity: 21%
Fold: GTP cyclohydrolase I feedback regulatory protein, GFRP
Superfamily: GTP cyclohydrolase I feedback regulatory protein, GFRP
Family: GTP cyclohydrolase I feedback regulatory protein, GFRP
Phyre2
| 18 |
|
PDB 2iea chain A domain 2
Region: 245 - 270
Aligned: 26
Modelled: 26
Confidence: 7.0%
Identity: 12%
Fold: Thiamin diphosphate-binding fold (THDP-binding)
Superfamily: Thiamin diphosphate-binding fold (THDP-binding)
Family: TK-like PP module
Phyre2
| 19 |
|
PDB 1ccw chain A
Region: 245 - 273
Aligned: 29
Modelled: 29
Confidence: 6.9%
Identity: 21%
Fold: Flavodoxin-like
Superfamily: Cobalamin (vitamin B12)-binding domain
Family: Cobalamin (vitamin B12)-binding domain
Phyre2
| 20 |
|
PDB 5vrv chain A
Region: 214 - 277
Aligned: 63
Modelled: 64
Confidence: 6.4%
Identity: 13%
PDB header:hydrolase,oxidoreductase
Chain: A: PDB Molecule:protein regulated by acid ph;
PDBTitle: 2.05 angstrom resolution crystal structure of c-terminal domain2 (duf2156) of putative lysylphosphatidylglycerol synthetase from3 agrobacterium fabrum.
Phyre2
| 21 |
|
| 22 |
|
| 23 |
|
| 24 |
|