| 1 |
|
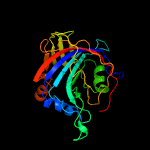
PDB 3bh2 chain C
Region: 3 - 236
Aligned: 215
Modelled: 234
Confidence: 100.0%
Identity: 20%
PDB header:lyase
Chain: C: PDB Molecule:acetoacetate decarboxylase;
PDBTitle: structural studies of acetoacetate decarboxylase
Phyre2
| 2 |
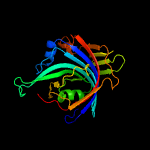
|
PDB 3bgt chain D
Region: 3 - 236
Aligned: 216
Modelled: 232
Confidence: 100.0%
Identity: 20%
PDB header:lyase
Chain: D: PDB Molecule:probable acetoacetate decarboxylase;
PDBTitle: structural studies of acetoacetate decarboxylase
Phyre2
| 3 |
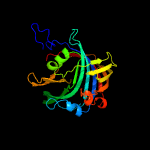
|
PDB 3c8w chain A domain 1
Region: 14 - 236
Aligned: 204
Modelled: 222
Confidence: 100.0%
Identity: 21%
Fold: Acetoacetate decarboxylase-like
Superfamily: Acetoacetate decarboxylase-like
Family: Acetoacetate decarboxylase-like
Phyre2
| 4 |
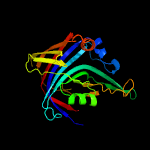
|
PDB 4jme chain A
Region: 12 - 236
Aligned: 202
Modelled: 225
Confidence: 100.0%
Identity: 19%
PDB header:unknown function
Chain: A: PDB Molecule:putative uncharacterized protein mppr;
PDBTitle: enduracididine biosynthesis enzyme mppr complexed with 2-keto-2 enduracididine
Phyre2
| 5 |
|
PDB 5upb chain A
Region: 11 - 236
Aligned: 201
Modelled: 221
Confidence: 100.0%
Identity: 17%
PDB header:lyase
Chain: A: PDB Molecule:acetoacetate decarboxylase;
PDBTitle: swit_4259, an acetoacetate decarboxylase-like enzyme from sphingomonas2 wittichii rw1
Phyre2
| 6 |
|
PDB 4zbt chain B
Region: 1 - 232
Aligned: 216
Modelled: 232
Confidence: 100.0%
Identity: 25%
PDB header:lyase
Chain: B: PDB Molecule:acetoacetate decarboxylase;
PDBTitle: streptomyces bingchenggensis aldolase-dehydratase in schiff base2 complex with pyruvate
Phyre2
| 7 |
|
PDB 3cmb chain A domain 1
Region: 14 - 234
Aligned: 209
Modelled: 218
Confidence: 99.9%
Identity: 16%
Fold: Acetoacetate decarboxylase-like
Superfamily: Acetoacetate decarboxylase-like
Family: Acetoacetate decarboxylase-like
Phyre2
| 8 |
|
PDB 1tg7 chain A domain 1
Region: 117 - 153
Aligned: 36
Modelled: 37
Confidence: 40.5%
Identity: 17%
Fold: Beta-galactosidase LacA, domain 3
Superfamily: Beta-galactosidase LacA, domain 3
Family: Beta-galactosidase LacA, domain 3
Phyre2
| 9 |
|
PDB 5tey chain B
Region: 101 - 130
Aligned: 29
Modelled: 30
Confidence: 17.7%
Identity: 24%
PDB header:transferase
Chain: B: PDB Molecule:n6-adenosine-methyltransferase subunit mettl14;
PDBTitle: human mettl3-mettl14 complex
Phyre2
| 10 |
|
PDB 2jsp chain A
Region: 116 - 123
Aligned: 8
Modelled: 8
Confidence: 6.5%
Identity: 38%
PDB header:gene regulation
Chain: A: PDB Molecule:transcriptional regulatory protein ros;
PDBTitle: the prokaryotic cys2his2 zinc finger adopts a novel fold as2 revealed by the nmr structure of a. tumefaciens ros dna3 binding domain
Phyre2
| 11 |
|
PDB 6nu9 chain A
Region: 200 - 233
Aligned: 32
Modelled: 34
Confidence: 6.2%
Identity: 19%
PDB header:viral protein
Chain: A: PDB Molecule:zinc-binding non-structural protein;
PDBTitle: crystal structure of a zinc-binding non-structural protein from the2 hepatitis e virus
Phyre2
| 12 |
|
PDB 1cf4 chain B
Region: 115 - 127
Aligned: 13
Modelled: 13
Confidence: 5.5%
Identity: 38%
PDB header:transferase
Chain: B: PDB Molecule:protein (activated p21cdc42hs kinase);
PDBTitle: cdc42/ack gtpase-binding domain complex
Phyre2
| 13 |
|
PDB 5mhj chain A
Region: 110 - 122
Aligned: 13
Modelled: 13
Confidence: 5.5%
Identity: 38%
PDB header:transcription
Chain: A: PDB Molecule:major viral transcription factor icp4;
PDBTitle: icp4 dna-binding domain, lacking intrinsically disordered region, in2 complex with 12mer dna duplex from its own promoter
Phyre2