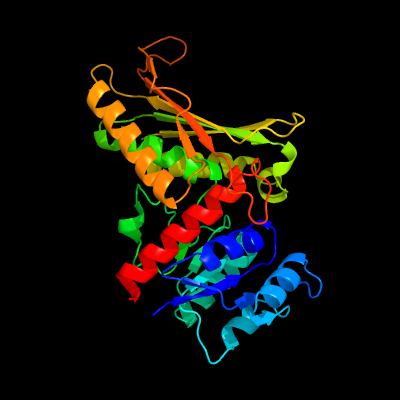
| 1 | c4jn6B_
|
|
 |
100.0 |
100 |
PDB header:lyase/oxidoreductase
Chain: B: PDB Molecule:acetaldehyde dehydrogenase;
PDBTitle: crystal structure of the aldolase-dehydrogenase complex from2 mycobacterium tuberculosis hrv37
|
|
|
|
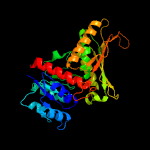
| 2 | c1nvmB_
|
|
 |
100.0 |
58 |
PDB header:lyase/oxidoreductase
Chain: B: PDB Molecule:acetaldehyde dehydrogenase (acylating);
PDBTitle: crystal structure of a bifunctional aldolase-dehydrogenase :2 sequestering a reactive and volatile intermediate
|
|
|
|
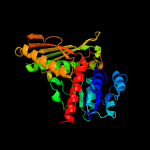
| 3 | d1nvmb1
|
|
 |
100.0 |
49 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
|
|
|
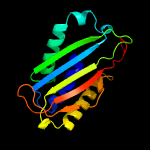
| 4 | d1nvmb2
|
|
 |
100.0 |
61 |
Fold:FwdE/GAPDH domain-like
Superfamily:Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domain
Family:GAPDH-like |
|
|
|
| 5 | c1b7gO_
|
|
 |
99.6 |
17 |
PDB header:oxidoreductase
Chain: O: PDB Molecule:protein (glyceraldehyde 3-phosphate dehydrogenase);
PDBTitle: glyceraldehyde 3-phosphate dehydrogenase
|
|
|
|
| 6 | c3kubA_
|
|
 |
99.6 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:aspartate-semialdehyde dehydrogenase;
PDBTitle: crystal structure of aspartate semi-aldehyde dehydrogenase complexed2 with glycerol and phosphate of mycobacterium tuberculosis h37rv
|
|
|
|
| 7 | c1cf2Q_
|
|
 |
99.5 |
16 |
PDB header:oxidoreductase
Chain: Q: PDB Molecule:protein (glyceraldehyde-3-phosphate dehydrogenase);
PDBTitle: three-dimensional structure of d-glyceraldehyde-3-phosphate2 dehydrogenase from the hyperthermophilic archaeon methanothermus3 fervidus
|
|
|
|
| 8 | c2qz9B_
|
|
 |
99.5 |
21 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:aspartate-semialdehyde dehydrogenase;
PDBTitle: crystal structure of aspartate semialdehyde dehydrogenase2 ii from vibrio cholerae
|
|
|
|
| 9 | d2hjsa1
|
|
 |
99.5 |
23 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
|
|
|
| 10 | c2czcD_
|
|
 |
99.5 |
17 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:glyceraldehyde-3-phosphate dehydrogenase;
PDBTitle: crystal structure of glyceraldehyde-3-phosphate dehydrogenase from2 pyrococcus horikoshii ot3
|
|
|
|
| 11 | c2hjsA_
|
|
 |
99.5 |
20 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:usg-1 protein homolog;
PDBTitle: the structure of a probable aspartate-semialdehyde dehydrogenase from2 pseudomonas aeruginosa
|
|
|
|
| 12 | c2ozpA_
|
|
 |
99.4 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:n-acetyl-gamma-glutamyl-phosphate reductase;
PDBTitle: crystal structure of n-acetyl-gamma-glutamyl-phosphate reductase2 (ttha1904) from thermus thermophilus
|
|
|
|
| 13 | d1t4ba1
|
|
 |
99.4 |
17 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
|
|
|
| 14 | d1cf2o1
|
|
 |
99.3 |
22 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
|
|
|
| 15 | c4dpkB_
|
|
 |
99.3 |
15 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:malonyl-coa/succinyl-coa reductase;
PDBTitle: structure of malonyl-coenzyme a reductase from crenarchaeota
|
|
|
|
| 16 | c2yv3B_
|
|
 |
99.3 |
19 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:aspartate-semialdehyde dehydrogenase;
PDBTitle: crystal structure of aspartate semialdehyde dehydrogenase from thermus2 thermophilus hb8
|
|
|
|
| 17 | c2yyyB_
|
|
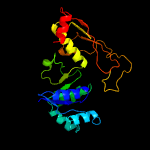 |
99.2 |
18 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:glyceraldehyde-3-phosphate dehydrogenase;
PDBTitle: crystal structure of glyceraldehyde-3-phosphate2 dehydrogenase
|
|
|
|
| 18 | c1ys4A_
|
|
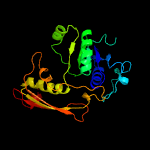 |
99.1 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:aspartate-semialdehyde dehydrogenase;
PDBTitle: structure of aspartate-semialdehyde dehydrogenase from methanococcus2 jannaschii
|
|
|
|
| 19 | c5jw6A_
|
|
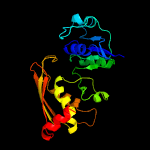 |
99.1 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:aspartate-semialdehyde dehydrogenase;
PDBTitle: cystal structure of aspartate semialdehyde dehydrogenase from2 aspergillus fumigatus
|
|
|
|
| 20 | c2gz3D_
|
|
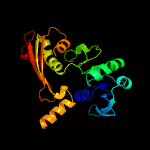 |
99.1 |
16 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:aspartate beta-semialdehyde dehydrogenase;
PDBTitle: structure of aspartate semialdehyde dehydrogenase (asadh) from2 streptococcus pneumoniae complexed with nadp and aspartate-3 semialdehyde
|
|
|
|
| 21 | c2ep5B_ |
|
not modelled |
99.1 |
14 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:350aa long hypothetical aspartate-semialdehyde
PDBTitle: structural study of project id st1242 from sulfolobus tokodaii strain7
|
|
|
| 22 | c1t4bB_ |
|
not modelled |
99.1 |
13 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:aspartate-semialdehyde dehydrogenase;
PDBTitle: 1.6 angstrom structure of esherichia coli aspartate-2 semialdehyde dehydrogenase.
|
|
|
| 23 | c5cefA_ |
|
not modelled |
99.0 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:aspartate-semialdehyde dehydrogenase;
PDBTitle: cystal structure of aspartate semialdehyde dehydrogenase from2 cryptococcus neoformans
|
|
|
| 24 | d2gz1a1 |
|
not modelled |
99.0 |
16 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
|
|
| 25 | c2g17A_ |
|
not modelled |
99.0 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:n-acetyl-gamma-glutamyl-phosphate reductase;
PDBTitle: the structure of n-acetyl-gamma-glutamyl-phosphate reductase from2 salmonella typhimurium.
|
|
|
| 26 | c1mb4B_ |
|
not modelled |
99.0 |
19 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:aspartate-semialdehyde dehydrogenase;
PDBTitle: crystal structure of aspartate semialdehyde dehydrogenase from vibrio2 cholerae with nadp and s-methyl-l-cysteine sulfoxide
|
|
|
| 27 | c2gd1P_ |
|
not modelled |
99.0 |
17 |
PDB header:oxidoreductase(aldehyde(d)-nad(a))
Chain: P: PDB Molecule:apo-d-glyceraldehyde-3-phosphate dehydrogenase;
PDBTitle: coenzyme-induced conformational changes in glyceraldehyde-3-2 phosphate dehydrogenase from bacillus stearothermophillus
|
|
|
| 28 | c4zhsC_ |
|
not modelled |
99.0 |
13 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:aspartate semialdehyde dehydrogenase;
PDBTitle: crystal structure of aspartate semialdehyde dehydrogenase from2 trichophyton rubrum
|
|
|
| 29 | c1s7cA_ |
|
not modelled |
99.0 |
23 |
PDB header:structural genomics, oxidoreductase
Chain: A: PDB Molecule:glyceraldehyde 3-phosphate dehydrogenase a;
PDBTitle: crystal structure of mes buffer bound form of glyceraldehyde 3-2 phosphate dehydrogenase from escherichia coli
|
|
|
| 30 | c2q49B_ |
|
not modelled |
98.9 |
12 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:probable n-acetyl-gamma-glutamyl-phosphate reductase;
PDBTitle: ensemble refinement of the protein crystal structure of gene product2 from arabidopsis thaliana at2g19940
|
|
|
| 31 | c4wojB_ |
|
not modelled |
98.9 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:aspartate semialdehyde dehydrogenase;
PDBTitle: aspartate semialdehyde dehydrogenase from francisella tularensis
|
|
|
| 32 | c3uw3A_ |
|
not modelled |
98.9 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:aspartate-semialdehyde dehydrogenase;
PDBTitle: crystal structure of an aspartate-semialdehyde dehydrogenase from2 burkholderia thailandensis
|
|
|
| 33 | c2i5pO_ |
|
not modelled |
98.8 |
19 |
PDB header:oxidoreductase
Chain: O: PDB Molecule:glyceraldehyde-3-phosphate dehydrogenase 1;
PDBTitle: crystal structure of glyceraldehyde-3-phosphate2 dehydrogenase isoform 1 from k. marxianus
|
|
|
| 34 | c3sthA_ |
|
not modelled |
98.8 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glyceraldehyde-3-phosphate dehydrogenase;
PDBTitle: crystal structure of glyceraldehyde-3-phosphate dehydrogenase from2 toxoplasma gondii
|
|
|
| 35 | c2d2iO_ |
|
not modelled |
98.8 |
19 |
PDB header:oxidoreductase
Chain: O: PDB Molecule:glyceraldehyde 3-phosphate dehydrogenase;
PDBTitle: crystal structure of nadp-dependent glyceraldehyde-3-2 phosphate dehydrogenase from synechococcus sp. complexed3 with nadp+
|
|
|
| 36 | c5ur0B_ |
|
not modelled |
98.8 |
18 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:glyceraldehyde-3-phosphate dehydrogenase;
PDBTitle: crystallographic structure of glyceraldehyde-3-phosphate dehydrogenase2 from naegleria gruberi
|
|
|
| 37 | c2pkrI_ |
|
not modelled |
98.7 |
20 |
PDB header:oxidoreductase
Chain: I: PDB Molecule:glyceraldehyde-3-phosphate dehydrogenase aor;
PDBTitle: crystal structure of (a+cte)4 chimeric form of photosyntetic2 glyceraldehyde-3-phosphate dehydrogenase, complexed with nadp
|
|
|
| 38 | c3h9eO_ |
|
not modelled |
98.7 |
18 |
PDB header:oxidoreductase
Chain: O: PDB Molecule:glyceraldehyde-3-phosphate dehydrogenase, testis-specific;
PDBTitle: crystal structure of human sperm-specific glyceraldehyde-3-phosphate2 dehydrogenase (gapds) complex with nad and phosphate
|
|
|
| 39 | c1hdgO_ |
|
not modelled |
98.7 |
19 |
PDB header:oxidoreductase (aldehy(d)-nad(a))
Chain: O: PDB Molecule:holo-d-glyceraldehyde-3-phosphate dehydrogenase;
PDBTitle: the crystal structure of holo-glyceraldehyde-3-phosphate dehydrogenase2 from the hyperthermophilic bacterium thermotoga maritima at 2.53 angstroms resolution
|
|
|
| 40 | c2x5kO_ |
|
not modelled |
98.6 |
16 |
PDB header:oxidoreductase
Chain: O: PDB Molecule:d-erythrose-4-phosphate dehydrogenase;
PDBTitle: structure of an active site mutant of the d-erythrose-4-phosphate2 dehydrogenase from e. coli
|
|
|
| 41 | c1rm4O_ |
|
not modelled |
98.6 |
20 |
PDB header:oxidoreductase
Chain: O: PDB Molecule:glyceraldehyde 3-phosphate dehydrogenase a;
PDBTitle: crystal structure of recombinant photosynthetic glyceraldehyde-3-2 phosphate dehydrogenase a4 isoform, complexed with nadp
|
|
|
| 42 | c4qx6A_ |
|
not modelled |
98.6 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glyceraldehyde 3-phosphate dehydrogenase;
PDBTitle: crystal structure of glyceraldehyde-3-phosphate dehydrogenase from2 streptococcus agalactiae nem316 at 2.46 angstrom resolution
|
|
|
| 43 | c3b20R_ |
|
not modelled |
98.6 |
18 |
PDB header:oxidoreductase
Chain: R: PDB Molecule:glyceraldehyde 3-phosphate dehydrogenase (nadp+);
PDBTitle: crystal structure of glyceraldehyde-3-phosphate dehydrogenase2 complexed with nadfrom synechococcus elongatus"
|
|
|
| 44 | c4dibF_ |
|
not modelled |
98.6 |
19 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:glyceraldehyde 3-phosphate dehydrogenase;
PDBTitle: the crystal structure of glyceraldehyde-3-phosphate dehydrogenase from2 bacillus anthracis str. sterne
|
|
|
| 45 | c6norB_ |
|
not modelled |
98.6 |
30 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative nad dependent dehydrogenase;
PDBTitle: crystal structure of gend2 from gentamicin a biosynthesis in complex2 with nad
|
|
|
| 46 | c2ep7B_ |
|
not modelled |
98.6 |
26 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:glyceraldehyde-3-phosphate dehydrogenase;
PDBTitle: structural study of project id aq_1065 from aquifex aeolicus vf5
|
|
|
| 47 | c5j9gB_ |
|
not modelled |
98.6 |
18 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:glyceraldehyde-3-p dehydrogenase;
PDBTitle: structure of lactobacillus acidophilus glyceraldehyde-3-phosphate2 dehydrogenase at 2.21 angstrom resolution
|
|
|
| 48 | c3e18A_ |
|
not modelled |
98.6 |
27 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:oxidoreductase;
PDBTitle: crystal structure of nad-binding protein from listeria innocua
|
|
|
| 49 | c5kt0A_ |
|
not modelled |
98.5 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:4-hydroxy-tetrahydrodipicolinate reductase;
PDBTitle: dihydrodipicolinate reductase from the industrial and evolutionarily2 important cyanobacteria anabaena variabilis.
|
|
|
| 50 | c3cieC_ |
|
not modelled |
98.5 |
17 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:glyceraldehyde-3-phosphate dehydrogenase;
PDBTitle: crystal structure of glyceraldehyde 3-phosphate2 dehydrogenase from cryptosporidium parvum
|
|
|
| 51 | c3docD_ |
|
not modelled |
98.5 |
17 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:glyceraldehyde 3-phosphate dehydrogenase;
PDBTitle: crystal structure of trka glyceraldehyde-3-phosphate dehydrogenase2 from brucella melitensis
|
|
|
| 52 | c3upyB_ |
|
not modelled |
98.5 |
21 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:oxidoreductase;
PDBTitle: crystal structure of the brucella abortus enzyme catalyzing the first2 committed step of the methylerythritol 4-phosphate pathway.
|
|
|
| 53 | c3wb9A_ |
|
not modelled |
98.5 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:diaminopimelate dehydrogenase;
PDBTitle: crystal structures of meso-diaminopimelate dehydrogenase from2 symbiobacterium thermophilum
|
|
|
| 54 | c4hadD_ |
|
not modelled |
98.5 |
18 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:probable oxidoreductase protein;
PDBTitle: crystal structure of probable oxidoreductase protein from rhizobium2 etli cfn 42
|
|
|
| 55 | c3wgzB_ |
|
not modelled |
98.5 |
14 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:meso-diaminopimelate dehydrogenase;
PDBTitle: crystal structure of meso-dapdh q154l/t173i/r199m/p248s/h249n/n276s2 mutant with d-leucine of from clostridium tetani e88
|
|
|
| 56 | d2czca2 |
|
not modelled |
98.5 |
29 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
|
|
| 57 | c5uibA_ |
|
not modelled |
98.4 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:oxidoreductase protein;
PDBTitle: crystal structure of an oxidoreductase from agrobacterium radiobacter2 in complex with nad+, l-tartaric acid and magnesium
|
|
|
| 58 | c2b4rQ_ |
|
not modelled |
98.4 |
15 |
PDB header:oxidoreductase
Chain: Q: PDB Molecule:glyceraldehyde-3-phosphate dehydrogenase;
PDBTitle: crystal structure of glyceraldehyde-3-phosphate dehydrogenase from2 plasmodium falciparum at 2.25 angstrom resolution reveals intriguing3 extra electron density in the active site
|
|
|
| 59 | c3wycB_ |
|
not modelled |
98.4 |
15 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:meso-diaminopimelate d-dehydrogenase;
PDBTitle: structure of a meso-diaminopimelate dehydrogenase in complex with nadp
|
|
|
| 60 | c6iauB_ |
|
not modelled |
98.4 |
18 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:amine dehydrogenase;
PDBTitle: amine dehydrogenase from cystobacter fuscus in complex with nadp+ and2 cyclohexylamine
|
|
|
| 61 | c1cerC_ |
|
not modelled |
98.4 |
18 |
PDB header:oxidoreductase (aldehyde(d)-nad(a))
Chain: C: PDB Molecule:holo-d-glyceraldehyde-3-phosphate dehydrogenase;
PDBTitle: determinants of enzyme thermostability observed in the2 molecular structure of thermus aquaticus d-glyceraldehyde-3 3-phosphate dehydrogenase at 2.5 angstroms resolution
|
|
|
| 62 | c6ok4A_ |
|
not modelled |
98.4 |
25 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glyceraldehyde-3-phosphate dehydrogenase;
PDBTitle: crystal structure of glyceraldehyde-3-phosphate dehydrogenase (gapdh)2 from chlamydia trachomatis with bound nad
|
|
|
| 63 | c3db2C_ |
|
not modelled |
98.4 |
16 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:putative nadph-dependent oxidoreductase;
PDBTitle: crystal structure of a putative nadph-dependent oxidoreductase2 (dhaf_2064) from desulfitobacterium hafniense dcb-2 at 1.70 a3 resolution
|
|
|
| 64 | c4mkzA_ |
|
not modelled |
98.4 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:inositol dehydrogenase;
PDBTitle: crystal structure of apo scyllo-inositol dehydrogenase from2 lactobacillus casei at 77k
|
|
|
| 65 | c2dc1A_ |
|
not modelled |
98.4 |
29 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:l-aspartate dehydrogenase;
PDBTitle: crystal structure of l-aspartate dehydrogenase from2 hyperthermophilic archaeon archaeoglobus fulgidus
|
|
|
| 66 | d2g17a1 |
|
not modelled |
98.4 |
23 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
|
|
| 67 | c5b3uB_ |
|
not modelled |
98.4 |
22 |
PDB header:transferase
Chain: B: PDB Molecule:biliverdin reductase;
PDBTitle: crystal structure of biliverdin reductase in complex with nadp+ from2 synechocystis sp. pcc 6803
|
|
|
| 68 | c3hq4R_ |
|
not modelled |
98.4 |
24 |
PDB header:oxidoreductase
Chain: R: PDB Molecule:glyceraldehyde-3-phosphate dehydrogenase 1;
PDBTitle: crystal structure of c151s mutant of glyceraldehyde-3-phosphate2 dehydrogenase 1 (gapdh1) complexed with nad from staphylococcus3 aureus mrsa252 at 2.2 angstrom resolution
|
|
|
| 69 | c3rbvA_ |
|
not modelled |
98.4 |
20 |
PDB header:sugar binding protein
Chain: A: PDB Molecule:sugar 3-ketoreductase;
PDBTitle: crystal structure of kijd10, a 3-ketoreductase from actinomadura2 kijaniata incomplex with nadp
|
|
|
| 70 | c3ceaA_ |
|
not modelled |
98.4 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:myo-inositol 2-dehydrogenase;
PDBTitle: crystal structure of myo-inositol 2-dehydrogenase (np_786804.1) from2 lactobacillus plantarum at 2.40 a resolution
|
|
|
| 71 | c1ihxD_ |
|
not modelled |
98.3 |
18 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:glyceraldehyde 3-phosphate dehydrogenase;
PDBTitle: crystal structure of two d-glyceraldehyde-3-phosphate2 dehydrogenase complexes: a case of asymmetry
|
|
|
| 72 | c3e9mC_ |
|
not modelled |
98.3 |
18 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:oxidoreductase, gfo/idh/moca family;
PDBTitle: crystal structure of an oxidoreductase from enterococcus2 faecalis
|
|
|
| 73 | c2q4eB_ |
|
not modelled |
98.3 |
19 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:probable oxidoreductase at4g09670;
PDBTitle: ensemble refinement of the protein crystal structure of gene product2 from arabidopsis thaliana at4g09670
|
|
|
| 74 | c4f3yA_ |
|
not modelled |
98.3 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dihydrodipicolinate reductase;
PDBTitle: x-ray crystal structure of dihydrodipicolinate reductase from2 burkholderia thailandensis
|
|
|
| 75 | c2glxD_ |
|
not modelled |
98.3 |
20 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:1,5-anhydro-d-fructose reductase;
PDBTitle: crystal structure analysis of bacterial 1,5-af reductase
|
|
|
| 76 | c1xeaD_ |
|
not modelled |
98.3 |
21 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:oxidoreductase, gfo/idh/moca family;
PDBTitle: crystal structure of a gfo/idh/moca family oxidoreductase2 from vibrio cholerae
|
|
|
| 77 | c2i3aD_ |
|
not modelled |
98.3 |
19 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:n-acetyl-gamma-glutamyl-phosphate reductase;
PDBTitle: crystal structure of n-acetyl-gamma-glutamyl-phosphate reductase2 (rv1652) from mycobacterium tuberculosis
|
|
|
| 78 | c1vknC_ |
|
not modelled |
98.3 |
17 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:n-acetyl-gamma-glutamyl-phosphate reductase;
PDBTitle: crystal structure of n-acetyl-gamma-glutamyl-phosphate reductase2 (tm1782) from thermotoga maritima at 1.80 a resolution
|
|
|
| 79 | c3hskB_ |
|
not modelled |
98.3 |
18 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:aspartate-semialdehyde dehydrogenase;
PDBTitle: crystal structure of aspartate semialdehyde dehydrogenase with nadp2 from candida albicans
|
|
|
| 80 | c3bioB_ |
|
not modelled |
98.3 |
23 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:oxidoreductase, gfo/idh/moca family;
PDBTitle: crystal structure of oxidoreductase (gfo/idh/moca family member) from2 porphyromonas gingivalis w83
|
|
|
| 81 | d2q49a1 |
|
not modelled |
98.3 |
9 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
|
|
| 82 | c3euwB_ |
|
not modelled |
98.3 |
22 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:myo-inositol dehydrogenase;
PDBTitle: crystal structure of a myo-inositol dehydrogenase from corynebacterium2 glutamicum atcc 13032
|
|
|
| 83 | c3ec7C_ |
|
not modelled |
98.3 |
23 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:putative dehydrogenase;
PDBTitle: crystal structure of putative dehydrogenase from salmonella2 typhimurium lt2
|
|
|
| 84 | c1h6dL_ |
|
not modelled |
98.2 |
19 |
PDB header:protein translocation
Chain: L: PDB Molecule:precursor form of glucose-fructose
PDBTitle: oxidized precursor form of glucose-fructose oxidoreductase2 from zymomonas mobilis complexed with glycerol
|
|
|
| 85 | c3dapB_ |
|
not modelled |
98.2 |
17 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:diaminopimelic acid dehydrogenase;
PDBTitle: c. glutamicum dap dehydrogenase in complex with nadp+ and2 the inhibitor 5s-isoxazoline
|
|
|
| 86 | c3evnA_ |
|
not modelled |
98.2 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:oxidoreductase, gfo/idh/moca family;
PDBTitle: crystal structure of putative oxidoreductase from streptococcus2 agalactiae 2603v/r
|
|
|
| 87 | c6iaqA_ |
|
not modelled |
98.2 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dihydrodipicolinate reductase n-terminus domain-containing
PDBTitle: structure of amine dehydrogenase from mycobacterium smegmatis
|
|
|
| 88 | c1drwA_ |
|
not modelled |
98.2 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dihydrodipicolinate reductase;
PDBTitle: escherichia coli dhpr/nhdh complex
|
|
|
| 89 | c1ofgF_ |
|
not modelled |
98.2 |
19 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:glucose-fructose oxidoreductase;
PDBTitle: glucose-fructose oxidoreductase
|
|
|
| 90 | c5ugjC_ |
|
not modelled |
98.2 |
20 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:4-hydroxy-tetrahydrodipicolinate reductase;
PDBTitle: crystal structure of htpa reductase from neisseria meningitidis
|
|
|
| 91 | d1vkna1 |
|
not modelled |
98.2 |
18 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
|
|
| 92 | c1evjC_ |
|
not modelled |
98.2 |
20 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:glucose-fructose oxidoreductase;
PDBTitle: crystal structure of glucose-fructose oxidoreductase (gfor)2 delta1-22 s64d
|
|
|
| 93 | c1obfO_ |
|
not modelled |
98.2 |
15 |
PDB header:glycolytic pathway
Chain: O: PDB Molecule:glyceraldehyde 3-phosphate dehydrogenase;
PDBTitle: the crystal structure of glyceraldehyde 3-phosphate2 dehydrogenase from alcaligenes xylosoxidans at 1.7 a3 resolution.
|
|
|
| 94 | c4ew6A_ |
|
not modelled |
98.2 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:d-galactose-1-dehydrogenase protein;
PDBTitle: crystal structure of d-galactose-1-dehydrogenase protein from2 rhizobium etli
|
|
|
| 95 | c3ezyB_ |
|
not modelled |
98.2 |
18 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:dehydrogenase;
PDBTitle: crystal structure of probable dehydrogenase tm_0414 from thermotoga2 maritima
|
|
|
| 96 | d1f06a1 |
|
not modelled |
98.2 |
16 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
|
|
| 97 | c3fd8A_ |
|
not modelled |
98.1 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:oxidoreductase, gfo/idh/moca family;
PDBTitle: crystal structure of an oxidoreductase from enterococcus2 faecalis
|
|
|
| 98 | c4gqaC_ |
|
not modelled |
98.1 |
20 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:nad binding oxidoreductase;
PDBTitle: crystal structure of nad binding oxidoreductase from klebsiella2 pneumoniae
|
|
|
| 99 | c4hktA_ |
|
not modelled |
98.1 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:inositol 2-dehydrogenase;
PDBTitle: crystal structure of a putative myo-inositol dehydrogenase from2 sinorhizobium meliloti 1021 (target psi-012312)
|
|
|
| 100 | c2o48X_ |
|
not modelled |
98.1 |
19 |
PDB header:oxidoreductase
Chain: X: PDB Molecule:dimeric dihydrodiol dehydrogenase;
PDBTitle: crystal structure of mammalian dimeric dihydrodiol dehydrogenase
|
|
|
| 101 | d1mb4a1 |
|
not modelled |
98.1 |
20 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
|
|
| 102 | d2dt5a2 |
|
not modelled |
98.1 |
22 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Transcriptional repressor Rex, C-terminal domain |
|
|
| 103 | c1zh8B_ |
|
not modelled |
98.1 |
19 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:oxidoreductase;
PDBTitle: crystal structure of oxidoreductase (tm0312) from thermotoga maritima2 at 2.50 a resolution
|
|
|
| 104 | c3gfgB_ |
|
not modelled |
98.1 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:uncharacterized oxidoreductase yvaa;
PDBTitle: structure of putative oxidoreductase yvaa from bacillus subtilis in2 triclinic form
|
|
|
| 105 | c6jnkA_ |
|
not modelled |
98.1 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:l-arabinose 1-dehydrogenase (nad(p)(+));
PDBTitle: crystal structure of azospirillum brasilense l-arabinose 1-2 dehydrogenase (nadp-bound form)
|
|
|
| 106 | c3q2kB_ |
|
not modelled |
98.1 |
24 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:oxidoreductase;
PDBTitle: crystal structure of the wlba dehydrogenase from bordetella pertussis2 in complex with nadh and udp-glcnaca
|
|
|
| 107 | c3dtyA_ |
|
not modelled |
98.1 |
25 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:oxidoreductase, gfo/idh/moca family;
PDBTitle: crystal structure of an oxidoreductase from pseudomonas2 syringae
|
|
|
| 108 | c3m2tA_ |
|
not modelled |
98.0 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:probable dehydrogenase;
PDBTitle: the crystal structure of dehydrogenase from chromobacterium2 violaceum
|
|
|
| 109 | d1dssg1 |
|
not modelled |
98.0 |
13 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
|
|
| 110 | c3kuxA_ |
|
not modelled |
98.0 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative oxidoreductase;
PDBTitle: structure of the ypo2259 putative oxidoreductase from yersinia pestis
|
|
|
| 111 | c2ho3D_ |
|
not modelled |
98.0 |
19 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:oxidoreductase, gfo/idh/moca family;
PDBTitle: crystal structure of oxidoreductase, gfo/idh/moca family from2 streptococcus pneumoniae
|
|
|
| 112 | c5a06E_ |
|
not modelled |
98.0 |
24 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:aldose-aldose oxidoreductase;
PDBTitle: crystal structure of aldose-aldose oxidoreductase from2 caulobacter crescentus complexed with sorbitol
|
|
|
| 113 | c2p2sA_ |
|
not modelled |
98.0 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative oxidoreductase;
PDBTitle: crystal structure of putative oxidoreductase (yp_050235.1) from2 erwinia carotovora atroseptica scri1043 at 1.25 a resolution
|
|
|
| 114 | c4h3vA_ |
|
not modelled |
98.0 |
26 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:oxidoreductase domain protein;
PDBTitle: crystal structure of oxidoreductase domain protein from kribbella2 flavida
|
|
|
| 115 | d2cvoa1 |
|
not modelled |
98.0 |
11 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
|
|
| 116 | c3nt5B_ |
|
not modelled |
98.0 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:inositol 2-dehydrogenase/d-chiro-inositol 3-dehydrogenase;
PDBTitle: crystal structure of myo-inositol dehydrogenase from bacillus subtilis2 with bound cofactor and product inosose
|
|
|
| 117 | c5yabD_ |
|
not modelled |
98.0 |
15 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:scyllo-inositol dehydrogenase with l-glucose dehydrogenase
PDBTitle: crystal structure of scyllo-inositol dehydrogenase with l-glucose2 dehydrogenase activity
|
|
|
| 118 | c5tenH_ |
|
not modelled |
98.0 |
12 |
PDB header:oxidoreductase
Chain: H: PDB Molecule:4-hydroxy-tetrahydrodipicolinate reductase;
PDBTitle: structure of 4-hydroxy-tetrahydrodipicolinate reductase from vibrio2 vulnificus with 2,5 furan dicarboxylic and nadh with intact3 polyhistidine tag
|
|
|
| 119 | c3hjaB_ |
|
not modelled |
97.9 |
20 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:glyceraldehyde-3-phosphate dehydrogenase;
PDBTitle: crystal structure of glyceraldehyde-3-phosphate dehydrogenase from2 borrelia burgdorferi
|
|
|
| 120 | c3ic5A_ |
|
not modelled |
97.9 |
19 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative saccharopine dehydrogenase;
PDBTitle: n-terminal domain of putative saccharopine dehydrogenase from ruegeria2 pomeroyi.
|
|
|