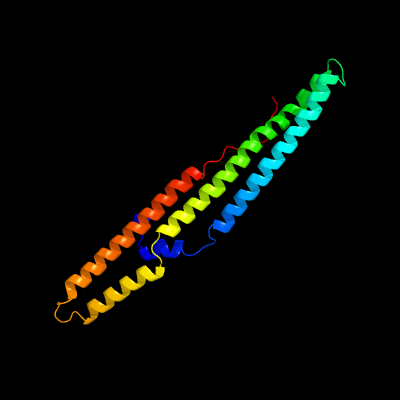
1 c5xfsB_
100.0
53
PDB header: protein transportChain: B: PDB Molecule: ppe family protein ppe15;PDBTitle: crystal structure of pe8-ppe15 in complex with espg5 from m.2 tuberculosis
2 c2g38B_
100.0
31
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: ppe family protein;PDBTitle: a pe/ppe protein complex from mycobacterium tuberculosis
3 d2g38b1
100.0
31
Fold: Ferritin-likeSuperfamily: PE/PPE dimer-likeFamily: PPE
4 c4xy3A_
100.0
17
PDB header: protein transportChain: A: PDB Molecule: esx-1 secretion-associated protein espb;PDBTitle: structure of esx-1 secreted protein espb
5 c4wj2A_
98.8
18
PDB header: unknown functionChain: A: PDB Molecule: antigen mtb48;PDBTitle: mycobacterial protein
6 c2vs0B_
98.1
8
PDB header: cell invasionChain: B: PDB Molecule: virulence factor esxa;PDBTitle: structural analysis of homodimeric staphylococcal aureus2 virulence factor esxa
7 c3gvmA_
97.9
11
PDB header: viral proteinChain: A: PDB Molecule: putative uncharacterized protein sag1039;PDBTitle: structure of the homodimeric wxg-100 family protein from streptococcus2 agalactiae
8 c4iogD_
97.8
13
PDB header: unknown functionChain: D: PDB Molecule: secreted protein esxb;PDBTitle: the crystal structure of a secreted protein esxb (wild-type, in p212 space group) from bacillus anthracis str. sterne
9 c3zbhC_
97.8
13
PDB header: unknown functionChain: C: PDB Molecule: esxa;PDBTitle: geobacillus thermodenitrificans esxa crystal form i
10 d1wa8a1
97.4
18
Fold: Ferritin-likeSuperfamily: EsxAB dimer-likeFamily: ESAT-6 like
11 c4lwsA_
96.7
19
PDB header: unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: esxa : esxb (semet) hetero-dimer from thermomonospora curvata
12 c4lwsB_
96.5
10
PDB header: unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: esxa : esxb (semet) hetero-dimer from thermomonospora curvata
13 d1wa8b1
96.3
19
Fold: Ferritin-likeSuperfamily: EsxAB dimer-likeFamily: ESAT-6 like
14 c4i0xA_
95.5
16
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: esat-6-like protein mab_3112;PDBTitle: crystal structure of the mycobacterum abscessus esxef (mab_3112-2 mab_3113) complex
15 c2kg7B_
93.6
19
PDB header: unknown functionChain: B: PDB Molecule: esat-6-like protein esxh;PDBTitle: structure and features of the complex formed by the tuberculosis2 virulence factors rv0287 and rv0288
16 c4i0xJ_
77.4
17
PDB header: structural genomics, unknown functionChain: J: PDB Molecule: esat-6-like protein mab_3113;PDBTitle: crystal structure of the mycobacterum abscessus esxef (mab_3112-2 mab_3113) complex
17 d1ui5a2
77.4
26
Fold: Tetracyclin repressor-like, C-terminal domainSuperfamily: Tetracyclin repressor-like, C-terminal domainFamily: Tetracyclin repressor-like, C-terminal domain
18 d1xkna_
51.6
17
Fold: Pentein, beta/alpha-propellerSuperfamily: PenteinFamily: Porphyromonas-type peptidylarginine deiminase
19 c3h6pB_
27.8
37
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: esat-6 like protein esxs;PDBTitle: crystal structure of rv3019c-rv3020c from mycobacterium tuberculosis
20 c3r5zB_
27.2
11
PDB header: unknown functionChain: B: PDB Molecule: putative uncharacterized protein;PDBTitle: structure of a deazaflavin-dependent reductase from nocardia2 farcinica, with co-factor f420
21 c2kg7A_
not modelled
27.0
39
PDB header: unknown functionChain: A: PDB Molecule: uncharacterized protein esxg (pe family protein);PDBTitle: structure and features of the complex formed by the tuberculosis2 virulence factors rv0287 and rv0288
22 c2iu1A_
not modelled
26.6
22
PDB header: transcriptionChain: A: PDB Molecule: eukaryotic translation initiation factor 5;PDBTitle: crystal structure of eif5 c-terminal domain
23 c4y9iA_
not modelled
26.3
18
PDB header: oxidoreductaseChain: A: PDB Molecule: mycobacterium tuberculosis paralogous family 11;PDBTitle: structure of f420-h2 dependent reductase (fdr-a) msmeg_2027
24 c4n91A_
not modelled
23.6
13
PDB header: transport proteinChain: A: PDB Molecule: trap dicarboxylate transporter, dctp subunit;PDBTitle: crystal structure of a trap periplasmic solute binding protein from2 anaerococcus prevotii dsm 20548 (apre_1383), target efi-510023, with3 bound alpha/beta d-glucuronate
25 c5frgA_
not modelled
23.2
75
PDB header: protein bindingChain: A: PDB Molecule: formin-binding protein 1-like;PDBTitle: the nmr structure of the cdc42-interacting region of toca1
26 c2lyyB_
not modelled
23.1
15
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: nmr structure of the protein nb7890a from shewanella sp
27 c3r5wO_
not modelled
21.3
23
PDB header: oxidoreductaseChain: O: PDB Molecule: deazaflavin-dependent nitroreductase;PDBTitle: structure of ddn, the deazaflavin-dependent nitroreductase from2 mycobacterium tuberculosis involved in bioreductive activation of pa-3 824, with co-factor f420
28 c3jywF_
not modelled
20.4
46
PDB header: ribosomeChain: F: PDB Molecule: 60s ribosomal protein l7(a);PDBTitle: structure of the 60s proteins for eukaryotic ribosome based on cryo-em2 map of thermomyces lanuginosus ribosome at 8.9a resolution
29 c3zfsA_
not modelled
20.0
16
PDB header: oxidoreductaseChain: A: PDB Molecule: f420-reducing hydrogenase, subunit alpha;PDBTitle: cryo-em structure of the f420-reducing nife-hydrogenase from a2 methanogenic archaeon with bound substrate
30 c1bkvA_
not modelled
20.0
38
PDB header: structural proteinChain: A: PDB Molecule: t3-785;PDBTitle: collagen
31 c2fulE_
not modelled
19.9
39
PDB header: translationChain: E: PDB Molecule: eukaryotic translation initiation factor 5;PDBTitle: crystal structure of the c-terminal domain of s. cerevisiae eif5
32 c2ahmG_
not modelled
19.0
12
PDB header: viral protein, replicationChain: G: PDB Molecule: replicase polyprotein 1ab, heavy chain;PDBTitle: crystal structure of sars-cov super complex of non-structural2 proteins: the hexadecamer
33 c1bkvC_
not modelled
18.7
38
PDB header: structural proteinChain: C: PDB Molecule: t3-785;PDBTitle: collagen
34 c1bkvB_
not modelled
18.7
38
PDB header: structural proteinChain: B: PDB Molecule: t3-785;PDBTitle: collagen
35 c3h96B_
not modelled
17.6
11
PDB header: flavoproteinChain: B: PDB Molecule: f420-h2 dependent reductase a;PDBTitle: msmeg_3358 f420 reductase
36 c2l5bA_
not modelled
17.5
41
PDB header: apoptosisChain: A: PDB Molecule: activator of apoptosis harakiri;PDBTitle: solution structure of the transmembrane domain of bcl-2 member2 harakiri in micelles
37 c2l5aA_
not modelled
17.2
22
PDB header: nuclear proteinChain: A: PDB Molecule: histone h3-like centromeric protein cse4, protein scm3,PDBTitle: structural basis for recognition of centromere specific histone h32 variant by nonhistone scm3
38 c2ke4A_
not modelled
16.0
75
PDB header: membrane proteinChain: A: PDB Molecule: cdc42-interacting protein 4;PDBTitle: the nmr structure of the tc10 and cdc42 interacting domain2 of cip4
39 c1paqA_
not modelled
16.0
15
PDB header: translationChain: A: PDB Molecule: translation initiation factor eif-2b epsilonPDBTitle: crystal structure of the catalytic fragment of eukaryotic2 initiation factor 2b epsilon
40 d1paqa_
not modelled
16.0
15
Fold: alpha-alpha superhelixSuperfamily: ARM repeatFamily: MIF4G domain-like
41 c4mnpA_
not modelled
16.0
16
PDB header: sugar binding proteinChain: A: PDB Molecule: n-acetylneuraminate-binding protein;PDBTitle: structure of the sialic acid binding protein from fusobacterium2 nucleatum subsp. nucleatum atcc 25586
42 c3r5yC_
not modelled
15.0
17
PDB header: unknown functionChain: C: PDB Molecule: putative uncharacterized protein;PDBTitle: structure of a deazaflavin-dependent nitroreductase from nocardia2 farcinica, with co-factor f420
43 c3ub0D_
not modelled
13.5
15
PDB header: replicationChain: D: PDB Molecule: non-structural protein 6, nsp6,;PDBTitle: crystal structure of the nonstructural protein 7 and 8 complex of2 feline coronavirus
44 c2kp7A_
not modelled
13.4
20
PDB header: hydrolaseChain: A: PDB Molecule: crossover junction endonuclease mus81;PDBTitle: solution nmr structure of the mus81 n-terminal hhh.2 northeast structural genomics consortium target mmt1a
45 c3juiA_
not modelled
13.3
15
PDB header: translationChain: A: PDB Molecule: translation initiation factor eif-2b subunit epsilon;PDBTitle: crystal structure of the c-terminal domain of human translation2 initiation factor eif2b epsilon subunit
46 c3trhI_
not modelled
13.2
19
PDB header: lyaseChain: I: PDB Molecule: phosphoribosylaminoimidazole carboxylasePDBTitle: structure of a phosphoribosylaminoimidazole carboxylase catalytic2 subunit (pure) from coxiella burnetii
47 d2ewoa1
not modelled
12.6
24
Fold: Pentein, beta/alpha-propellerSuperfamily: PenteinFamily: Porphyromonas-type peptidylarginine deiminase
48 c5lzkB_
not modelled
12.2
4
PDB header: structural genomicsChain: B: PDB Molecule: protein fam83b;PDBTitle: structure of the domain of unknown function duf1669 from human fam83b
49 c3qthA_
not modelled
11.6
9
PDB header: unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a dinb-like protein (cps_3021) from colwellia2 psychrerythraea 34h at 2.20 a resolution
50 c2hzkB_
not modelled
11.1
15
PDB header: ligand binding, transport proteinChain: B: PDB Molecule: trap-t family sorbitol/mannitol transporter, periplasmicPDBTitle: crystal structures of a sodium-alpha-keto acid binding subunit from a2 trap transporter in its open form
51 c2jerG_
not modelled
10.6
28
PDB header: hydrolaseChain: G: PDB Molecule: agmatine deiminase;PDBTitle: agmatine deiminase of enterococcus faecalis catalyzing its2 reaction.
52 c4el8A_
not modelled
10.2
31
PDB header: hydrolaseChain: A: PDB Molecule: glycoside hydrolase family 48;PDBTitle: the unliganded structure of c.bescii cela gh48 module
53 c2o01H_
not modelled
9.7
22
PDB header: photosynthesisChain: H: PDB Molecule: photosystem i reaction center subunit vi, chloroplast;PDBTitle: the structure of a plant photosystem i supercomplex at 3.4 angstrom2 resolution
54 c6b2wB_
not modelled
9.7
13
PDB header: hydrolaseChain: B: PDB Molecule: putative peptidyl-arginine deiminase family protein;PDBTitle: c. jejuni c315s agmatine deiminase with substrate bound
55 d2jera1
not modelled
9.5
28
Fold: Pentein, beta/alpha-propellerSuperfamily: PenteinFamily: Porphyromonas-type peptidylarginine deiminase
56 d1vkpa_
not modelled
9.5
31
Fold: Pentein, beta/alpha-propellerSuperfamily: PenteinFamily: Porphyromonas-type peptidylarginine deiminase
57 c3j3bF_
not modelled
9.1
31
PDB header: ribosomeChain: F: PDB Molecule: 60s ribosomal protein l7;PDBTitle: structure of the human 60s ribosomal proteins
58 c5ec0A_
not modelled
9.1
33
PDB header: structural proteinChain: A: PDB Molecule: alp7a;PDBTitle: crystal structure of actin-like protein alp7a
59 c3n6xA_
not modelled
9.1
13
PDB header: ligaseChain: A: PDB Molecule: putative glutathionylspermidine synthase;PDBTitle: crystal structure of a putative glutathionylspermidine synthase2 (mfla_0391) from methylobacillus flagellatus kt at 2.35 a resolution
60 c3j21Y_
not modelled
9.0
25
PDB header: ribosomeChain: Y: PDB Molecule: 50s ribosomal protein l30p;PDBTitle: promiscuous behavior of proteins in archaeal ribosomes revealed by2 cryo-em: implications for evolution of eukaryotic ribosomes (50s3 ribosomal proteins)
61 c5hl8B_
not modelled
9.0
18
PDB header: protein transportChain: B: PDB Molecule: type ii secretion system protein l;PDBTitle: 1.93 angstrom resolution crystal structure of a pullulanase-specific2 type ii secretion system integral cytoplasmic membrane protein gspl3 (c-terminal fragment; residues 309-397) from klebsiella pneumoniae4 subsp. pneumoniae ntuh-k2044
62 c4kkkA_
not modelled
8.8
19
PDB header: hydrolaseChain: A: PDB Molecule: exoglucanase s;PDBTitle: complex structure of catalytic domain of clostridium cellulovorans2 exgs and cellotetraose
63 c3b50A_
not modelled
8.7
14
PDB header: transport proteinChain: A: PDB Molecule: sialic acid-binding periplasmic protein siap;PDBTitle: structure of h. influenzae sialic acid binding protein2 bound to neu5ac.
64 c5i4rA_
not modelled
8.6
43
PDB header: toxin/antitoxinChain: A: PDB Molecule: contact-dependent inhibitor a;PDBTitle: contact-dependent inhibition system from escherichia coli nc101 -2 ternary cdia/cdii/ef-tu complex (trypsin-modified)
65 c1l2aD_
not modelled
8.5
19
PDB header: hydrolaseChain: D: PDB Molecule: cellobiohydrolase;PDBTitle: the crystal structure and catalytic mechanism of2 cellobiohydrolase cels, the major enzymatic component of3 the clostridium thermocellum cellulosome
66 d1l1ya_
not modelled
8.5
19
Fold: alpha/alpha toroidSuperfamily: Six-hairpin glycosidasesFamily: Cellulases catalytic domain
67 c4fusA_
not modelled
8.2
31
PDB header: hydrolaseChain: A: PDB Molecule: rtx toxins and related ca2+-binding protein;PDBTitle: the x-ray structure of hahella chejuensis family 48 glycosyl hydrolase
68 c2np3A_
not modelled
8.2
10
PDB header: transcriptionChain: A: PDB Molecule: putative tetr-family regulator;PDBTitle: crystal structure of tetr-family regulator (sco0857) from streptomyces2 coelicolor a3.
69 d2apla1
not modelled
8.2
9
Fold: PG0816-likeSuperfamily: PG0816-likeFamily: PG0816-like
70 c2nvjA_
not modelled
8.0
30
PDB header: hydrolaseChain: A: PDB Molecule: 25mer peptide from vacuolar atp synthase subunitPDBTitle: nmr structures of transmembrane segment from subunit a from2 the yeast proton v-atpase
71 c4i6jB_
not modelled
7.8
22
PDB header: transcriptionChain: B: PDB Molecule: f-box/lrr-repeat protein 3;PDBTitle: a ubiquitin ligase-substrate complex
72 c3j39F_
not modelled
7.7
31
PDB header: ribosomeChain: F: PDB Molecule: 60s ribosomal protein l7;PDBTitle: structure of the d. melanogaster 60s ribosomal proteins
73 c4jjjA_
not modelled
7.7
19
PDB header: hydrolaseChain: A: PDB Molecule: cellulose 1,4-beta-cellobiosidase;PDBTitle: the structure of t. fusca gh48 d224n mutant
74 c2kwuA_
not modelled
7.7
38
PDB header: protein binding/signaling proteinChain: A: PDB Molecule: dna polymerase iota;PDBTitle: solution structure of ubm2 of murine polymerase iota in complex with2 ubiquitin
75 c6o9l6_
not modelled
7.5
40
PDB header: transcription/dnaChain: 6: PDB Molecule: general transcription factor iih subunit 2;PDBTitle: human holo-pic in the closed state
76 c3fy6A_
not modelled
7.5
25
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: integron cassette protein;PDBTitle: structure from the mobile metagenome of v. cholerae. integron cassette2 protein vch_cass3
77 c5vmoB_
not modelled
7.5
50
PDB header: viral protein/apoptosisChain: B: PDB Molecule: bcl-2 interacting mediator of cell death;PDBTitle: crystal structure of grouper iridovirus giv66:bim complex
78 d2fgga1
not modelled
7.2
40
Fold: dsRBD-likeSuperfamily: Rv2632c-likeFamily: Rv2632c-like
79 c2jtwA_
not modelled
7.2
38
PDB header: membrane proteinChain: A: PDB Molecule: transmembrane helix 7 of yeast vatpase;PDBTitle: solution structure of tm7 bound to dpc micelles
80 c1bzgA_
not modelled
7.2
0
PDB header: hormoneChain: A: PDB Molecule: parathyroid hormone-related protein;PDBTitle: the solution structure of human parathyroid hormone-related2 protein (1-34) in near-physiological solution, nmr, 303 structures
81 c6epiC_
not modelled
7.2
20
PDB header: toxinChain: C: PDB Molecule: epsilon_1 antitoxin;PDBTitle: structure of the epsilon_1 / zeta_1 antitoxin / toxin system from2 neisseria gonorrhoeae in complex with unam-4p.
82 c3izce_
not modelled
7.1
19
PDB header: ribosomeChain: E: PDB Molecule: 60s ribosomal protein rpl11 (l5p);PDBTitle: localization of the large subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
83 c4c5eG_
not modelled
7.0
22
PDB header: transcriptionChain: G: PDB Molecule: polycomb protein pho;PDBTitle: crystal structure of the minimal pho-sfmbt complex (p21 spacegroup)
84 c4c5eH_
not modelled
7.0
22
PDB header: transcriptionChain: H: PDB Molecule: polycomb protein pho;PDBTitle: crystal structure of the minimal pho-sfmbt complex (p21 spacegroup)
85 c4xb6D_
not modelled
6.9
11
PDB header: transferaseChain: D: PDB Molecule: alpha-d-ribose 1-methylphosphonate 5-phosphate c-p lyase;PDBTitle: structure of the e. coli c-p lyase core complex
86 c1d0rA_
not modelled
6.9
30
PDB header: hormone/growth factorChain: A: PDB Molecule: glucagon-like peptide-1-(7-36)-amide;PDBTitle: solution structure of glucagon-like peptide-1-(7-36)-amide in2 trifluoroethanol/water
87 c6igzH_
not modelled
6.9
12
PDB header: plant proteinChain: H: PDB Molecule: psah;PDBTitle: structure of psi-lhci
88 d1kx5a_
not modelled
6.8
18
Fold: Histone-foldSuperfamily: Histone-foldFamily: Nucleosome core histones
89 c2yfwC_
not modelled
6.7
22
PDB header: cell cycleChain: C: PDB Molecule: histone h3-like centromeric protein cse4;PDBTitle: heterotetramer structure of kluyveromyces lactis cse4,h4
90 c6hu9u_
not modelled
6.7
27
PDB header: oxidoreductase/electron transportChain: U: PDB Molecule: cytochrome b-c1 complex subunit 10;PDBTitle: iii2-iv2 mitochondrial respiratory supercomplex from s. cerevisiae
91 c3a9rA_
not modelled
6.6
14
PDB header: isomeraseChain: A: PDB Molecule: d-arabinose isomerase;PDBTitle: x-ray structures of bacillus pallidus d-arabinose2 isomerasecomplex with (4r)-2-methylpentane-2,4-diol
92 c4wpyA_
not modelled
6.5
20
PDB header: de novo proteinChain: A: PDB Molecule: protein dl-rv1738;PDBTitle: racemic crystal structure of rv1738 from mycobacterium tuberculosis2 (form-ii)
93 c3zf7w_
not modelled
6.5
25
PDB header: ribosomeChain: W: PDB Molecule: 60s ribosomal protein l23, putative;PDBTitle: high-resolution cryo-electron microscopy structure of the trypanosoma2 brucei ribosome
94 c2zdjA_
not modelled
6.4
50
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein ttma177;PDBTitle: crystal structure of ttma177, a hypothetical protein from2 thermus thermophilus phage tma
95 d1g9ga_
not modelled
6.3
21
Fold: alpha/alpha toroidSuperfamily: Six-hairpin glycosidasesFamily: Cellulases catalytic domain
96 c4u0zH_
not modelled
6.3
12
PDB header: transferaseChain: H: PDB Molecule: adenosine monophosphate-protein transferase ficd;PDBTitle: eukaryotic fic domain containing protein with bound apcpp
97 d1vqow1
not modelled
6.3
25
Fold: Ribosomal protein L30p/L7eSuperfamily: Ribosomal protein L30p/L7eFamily: Ribosomal protein L30p/L7e
98 c3gyyC_
not modelled
6.2
13
PDB header: transport proteinChain: C: PDB Molecule: periplasmic substrate binding protein;PDBTitle: the ectoine binding protein of the teaabc trap transporter teaa in the2 apo-state
99 d1uadc_
not modelled
6.1
24
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: Other IPT/TIG domains