| 1 |
|
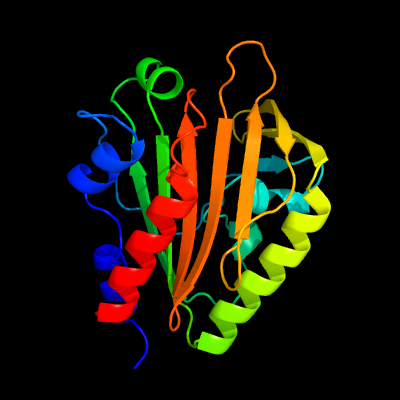
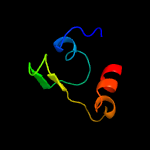
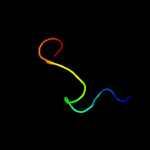
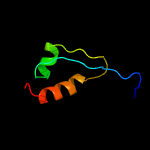
PDB 4esq chain A
Region: 41 - 237
Aligned: 192
Modelled: 197
Confidence: 100.0%
Identity: 34%
PDB header:transferase
Chain: A: PDB Molecule:serine/threonine protein kinase;
PDBTitle: crystal structure of the extracellular domain of pknh from2 mycobacterium tuberculosis
Phyre2
| 2 |
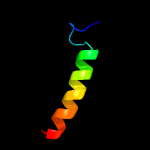
|
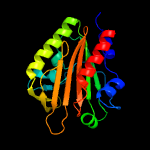
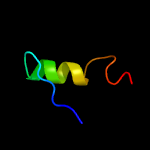
PDB 2b3g chain B
Region: 41 - 59
Aligned: 19
Modelled: 19
Confidence: 42.4%
Identity: 32%
PDB header:replication
Chain: B: PDB Molecule:cellular tumor antigen p53;
PDBTitle: p53n (fragment 33-60) bound to rpa70n
Phyre2
| 3 |
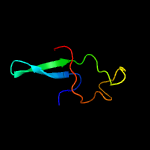
|
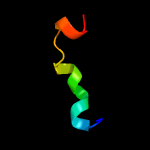
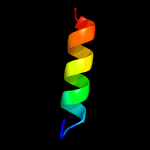
PDB 2fiu chain A domain 1
Region: 130 - 146
Aligned: 17
Modelled: 17
Confidence: 40.2%
Identity: 35%
Fold: Ferredoxin-like
Superfamily: Dimeric alpha+beta barrel
Family: Atu0297-like
Phyre2
| 4 |
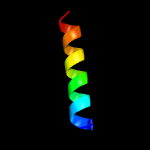
|
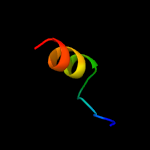
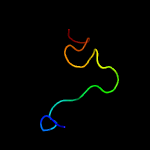
PDB 3lo3 chain E
Region: 130 - 146
Aligned: 17
Modelled: 17
Confidence: 40.2%
Identity: 35%
PDB header:structure genomics, unknown function
Chain: E: PDB Molecule:uncharacterized conserved protein;
PDBTitle: the crystal structure of a conserved functionally unknown protein from2 colwellia psychrerythraea 34h.
Phyre2
| 5 |
|
PDB 1jjc chain B domain 1
Region: 50 - 97
Aligned: 48
Modelled: 48
Confidence: 32.1%
Identity: 17%
Fold: Putative DNA-binding domain
Superfamily: Putative DNA-binding domain
Family: Domains B1 and B5 of PheRS-beta, PheT
Phyre2
| 6 |
|
PDB 2l14 chain B
Region: 41 - 59
Aligned: 19
Modelled: 19
Confidence: 30.1%
Identity: 32%
PDB header:protein binding
Chain: B: PDB Molecule:cellular tumor antigen p53;
PDBTitle: structure of cbp nuclear coactivator binding domain in complex with2 p53 tad
Phyre2
| 7 |
|
PDB 5uz5 chain C
Region: 129 - 152
Aligned: 24
Modelled: 24
Confidence: 27.6%
Identity: 25%
PDB header:nuclear protein/rna
Chain: C: PDB Molecule:u1 small nuclear ribonucleoprotein a,tap tag;
PDBTitle: s. cerevisiae u1 snrnp
Phyre2
| 8 |
|
PDB 3i08 chain D
Region: 134 - 150
Aligned: 17
Modelled: 17
Confidence: 23.6%
Identity: 24%
PDB header:signaling protein
Chain: D: PDB Molecule:neurogenic locus notch homolog protein 1;
PDBTitle: crystal structure of the s1-cleaved notch1 negative2 regulatory region (nrr)
Phyre2
| 9 |
|
PDB 2ly4 chain B
Region: 35 - 58
Aligned: 24
Modelled: 24
Confidence: 21.9%
Identity: 25%
PDB header:nuclear protein/antitumour protein
Chain: B: PDB Molecule:cellular tumor antigen p53;
PDBTitle: hmgb1-facilitated p53 dna binding occurs via hmg-box/p532 transactivation domain interaction and is regulated by the acidic3 tail
Phyre2
| 10 |
|
PDB 3dca chain C
Region: 130 - 146
Aligned: 17
Modelled: 13
Confidence: 19.2%
Identity: 12%
PDB header:structural genomics, unknown function
Chain: C: PDB Molecule:rpa0582;
PDBTitle: crystal structure of the rpa0582- protein of unknown2 function from rhodopseudomonas palustris- a structural3 genomics target
Phyre2
| 11 |
|
PDB 2mzd chain B
Region: 43 - 59
Aligned: 17
Modelled: 17
Confidence: 11.1%
Identity: 29%
PDB header:protein binding
Chain: B: PDB Molecule:cellular tumor antigen p53;
PDBTitle: characterization of the p300 taz2-p53 tad2 complex and comparison with2 the p300 taz2-p53 tad1 complex
Phyre2
| 12 |
|
PDB 6ehi chain I
Region: 129 - 154
Aligned: 26
Modelled: 26
Confidence: 9.7%
Identity: 12%
PDB header:dna binding protein
Chain: I: PDB Molecule:nuclease nuct;
PDBTitle: nuct from helicobacter pylori
Phyre2
| 13 |
|
PDB 2jp3 chain A
Region: 2 - 20
Aligned: 19
Modelled: 19
Confidence: 9.2%
Identity: 21%
PDB header:transcription
Chain: A: PDB Molecule:fxyd domain-containing ion transport regulator 4;
PDBTitle: solution structure of the human fxyd4 (chif) protein in sds2 micelles
Phyre2
| 14 |
|
PDB 2n8f chain A
Region: 150 - 157
Aligned: 8
Modelled: 8
Confidence: 9.1%
Identity: 38%
PDB header:toxin
Chain: A: PDB Molecule:spider toxin pi-hexatoxin-hi1a;
PDBTitle: chemical shift assignments and structure calculation of spider toxin2 pi-hexatoxin-hi1a
Phyre2
| 15 |
|
PDB 2ahm chain G
Region: 152 - 200
Aligned: 44
Modelled: 49
Confidence: 8.0%
Identity: 25%
PDB header:viral protein, replication
Chain: G: PDB Molecule:replicase polyprotein 1ab, heavy chain;
PDBTitle: crystal structure of sars-cov super complex of non-structural2 proteins: the hexadecamer
Phyre2
| 16 |
|
PDB 2jo1 chain A
Region: 2 - 20
Aligned: 19
Modelled: 19
Confidence: 7.5%
Identity: 16%
PDB header:hydrolase regulator
Chain: A: PDB Molecule:phospholemman;
PDBTitle: structure of the na,k-atpase regulatory protein fxyd1 in2 micelles
Phyre2
| 17 |
|
PDB 2mkv chain A
Region: 2 - 20
Aligned: 19
Modelled: 19
Confidence: 7.1%
Identity: 37%
PDB header:transport protein
Chain: A: PDB Molecule:sodium/potassium-transporting atpase subunit gamma;
PDBTitle: structure of the na,k-atpase regulatory protein fxyd2b in micelles
Phyre2
| 18 |
|
PDB 2dmw chain A
Region: 189 - 237
Aligned: 45
Modelled: 49
Confidence: 7.0%
Identity: 24%
PDB header:membrane protein
Chain: A: PDB Molecule:synaptobrevin-like 1 variant;
PDBTitle: solution structure of the longin domain of synaptobrevin-2 like protein 1
Phyre2
| 19 |
|
PDB 2zxe chain G
Region: 2 - 20
Aligned: 19
Modelled: 19
Confidence: 6.5%
Identity: 37%
PDB header:hydrolase/transport protein
Chain: G: PDB Molecule:phospholemman-like protein;
PDBTitle: crystal structure of the sodium - potassium pump in the e2.2k+.pi2 state
Phyre2
| 20 |
|
PDB 1xkm chain D
Region: 139 - 154
Aligned: 16
Modelled: 16
Confidence: 6.0%
Identity: 13%
PDB header:antibiotic
Chain: D: PDB Molecule:distinctin chain b;
PDBTitle: nmr structure of antimicrobial peptide distinctin in water
Phyre2
| 21 |
|
| 22 |
|
| 23 |
|
| 24 |
|