| 1 |
|
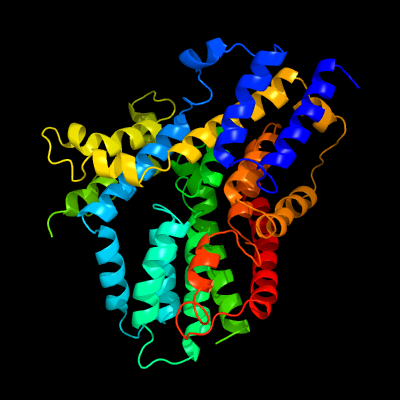
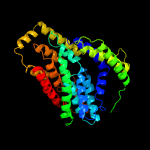
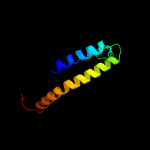
PDB 4f35 chain C
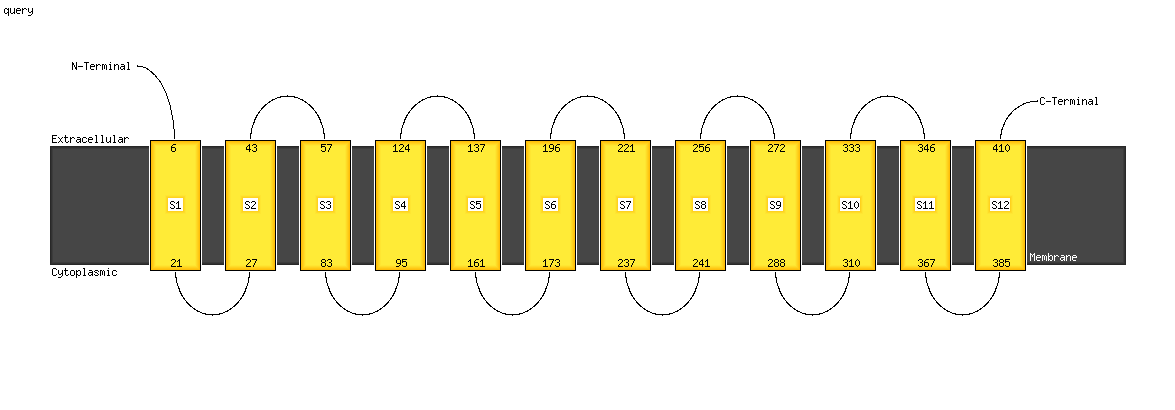
Region: 1 - 409
Aligned: 391
Modelled: 408
Confidence: 100.0%
Identity: 14%
PDB header:transport protein
Chain: C: PDB Molecule:transporter, nadc family;
PDBTitle: crystal structure of a bacterial dicarboxylate/sodium symporter
Phyre2
| 2 |
|
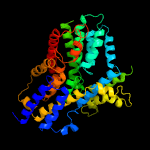
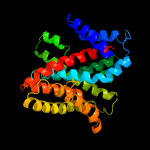
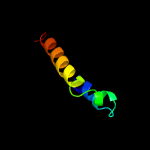
PDB 4r1i chain B
Region: 55 - 413
Aligned: 355
Modelled: 359
Confidence: 100.0%
Identity: 13%
PDB header:membrane protein
Chain: B: PDB Molecule:aminobenzoyl-glutamate transporter;
PDBTitle: structure and function of neisseria gonorrhoeae mtrf illuminates a2 class of antimetabolite efflux pumps
Phyre2
| 3 |
|
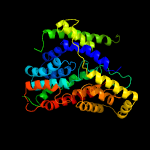
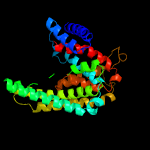
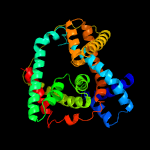
PDB 4r0c chain B
Region: 57 - 410
Aligned: 344
Modelled: 354
Confidence: 99.8%
Identity: 15%
PDB header:membrane protein
Chain: B: PDB Molecule:abgt putative transporter family;
PDBTitle: crystal structure of the alcanivorax borkumensis ydah transporter2 reveals an unusual topology
Phyre2
| 4 |
|
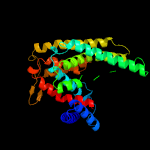
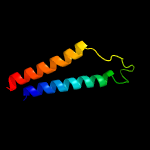
PDB 4czb chain B
Region: 3 - 287
Aligned: 279
Modelled: 285
Confidence: 69.7%
Identity: 10%
PDB header:membrane protein
Chain: B: PDB Molecule:na(+)/h(+) antiporter 1;
PDBTitle: structure of the sodium proton antiporter mjnhap1 from2 methanocaldococcus jannaschii at ph 8.
Phyre2
| 5 |
|
PDB 4bwz chain A
Region: 3 - 281
Aligned: 263
Modelled: 279
Confidence: 43.1%
Identity: 17%
PDB header:transport protein
Chain: A: PDB Molecule:na(+)/h(+) antiporter;
PDBTitle: crystal structure of the sodium proton antiporter, napa
Phyre2
| 6 |
|
PDB 5bz3 chain A
Region: 3 - 281
Aligned: 263
Modelled: 279
Confidence: 43.1%
Identity: 17%
PDB header:transport protein
Chain: A: PDB Molecule:na(+)/h(+) antiporter;
PDBTitle: crystal structure of sodium proton antiporter napa in outward-facing2 conformation.
Phyre2
| 7 |
|
PDB 3qnq chain D
Region: 135 - 210
Aligned: 76
Modelled: 76
Confidence: 23.7%
Identity: 11%
PDB header:membrane protein, transport protein
Chain: D: PDB Molecule:pts system, cellobiose-specific iic component;
PDBTitle: crystal structure of the transporter chbc, the iic component from the2 n,n'-diacetylchitobiose-specific phosphotransferase system
Phyre2
| 8 |
|
PDB 2bbj chain B
Region: 158 - 200
Aligned: 40
Modelled: 43
Confidence: 16.7%
Identity: 15%
PDB header:metal transport/membrane protein
Chain: B: PDB Molecule:divalent cation transport-related protein;
PDBTitle: crystal structure of the cora mg2+ transporter
Phyre2
| 9 |
|
PDB 5a1s chain B
Region: 2 - 324
Aligned: 298
Modelled: 308
Confidence: 11.9%
Identity: 14%
PDB header:transport protein
Chain: B: PDB Molecule:citrate-sodium symporter;
PDBTitle: crystal structure of the sodium-dependent citrate symporter secits2 form salmonella enterica.
Phyre2
| 10 |
|
PDB 1ar1 chain B
Region: 173 - 235
Aligned: 63
Modelled: 63
Confidence: 8.9%
Identity: 8%
PDB header:complex (oxidoreductase/antibody)
Chain: B: PDB Molecule:cytochrome c oxidase;
PDBTitle: structure at 2.7 angstrom resolution of the paracoccus2 denitrificans two-subunit cytochrome c oxidase complexed3 with an antibody fv fragment
Phyre2
| 11 |
|
PDB 1qle chain B
Region: 173 - 235
Aligned: 63
Modelled: 63
Confidence: 8.9%
Identity: 8%
PDB header:oxidoreductase/immune system
Chain: B: PDB Molecule:cytochrome c oxidase polypeptide ii;
PDBTitle: cryo-structure of the paracoccus denitrificans four-subunit cytochrome2 c oxidase in the completely oxidized state complexed with an antibody3 fv fragment
Phyre2
| 12 |
|
PDB 1m57 chain H
Region: 173 - 235
Aligned: 63
Modelled: 63
Confidence: 7.9%
Identity: 5%
PDB header:oxidoreductase
Chain: H: PDB Molecule:cytochrome c oxidase;
PDBTitle: structure of cytochrome c oxidase from rhodobacter2 sphaeroides (eq(i-286) mutant))
Phyre2
| 13 |
|
PDB 1fft chain B domain 2
Region: 169 - 235
Aligned: 67
Modelled: 67
Confidence: 7.2%
Identity: 15%
Fold: Transmembrane helix hairpin
Superfamily: Cytochrome c oxidase subunit II-like, transmembrane region
Family: Cytochrome c oxidase subunit II-like, transmembrane region
Phyre2
| 14 |
|
PDB 2knc chain A
Region: 173 - 211
Aligned: 39
Modelled: 39
Confidence: 6.1%
Identity: 15%
PDB header:cell adhesion
Chain: A: PDB Molecule:integrin alpha-iib;
PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
Phyre2
| 15 |
|
PDB 4us3 chain A
Region: 98 - 235
Aligned: 138
Modelled: 138
Confidence: 5.4%
Identity: 11%
PDB header:transport protein
Chain: A: PDB Molecule:transporter;
PDBTitle: crystal structure of the bacterial nss member mhst in an2 occluded inward-facing state
Phyre2