| 1 | c2xwlB_
|
|
 |
100.0 |
61 |
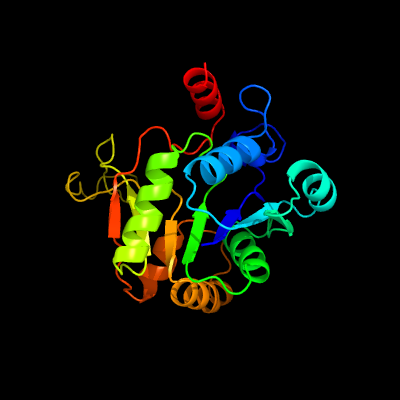
PDB header:transferase
Chain: B: PDB Molecule:2-c-methyl-d-erythritol 4-phosphate cytidylyltransferase;
PDBTitle: crystal structure of ispd from mycobacterium smegmatis in complex with2 ctp and mg
|
|
|
|
| 2 | d1i52a_
|
|
 |
100.0 |
29 |
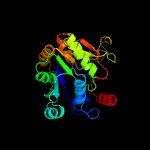
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:Cytidylytransferase |
|
|
|
| 3 | c5ddtA_
|
|
 |
100.0 |
25 |
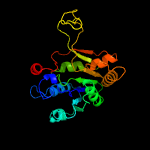
PDB header:transferase
Chain: A: PDB Molecule:2-c-methyl-d-erythritol 4-phosphate cytidylyltransferase;
PDBTitle: crystal structure of ispd from bacillus subtilis at 1.80 angstroms2 resolution, crystal form i
|
|
|
|
| 4 | d1vpaa_
|
|
 |
100.0 |
30 |
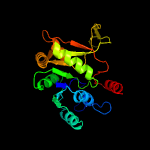
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:Cytidylytransferase |
|
|
|
| 5 | c4mybA_
|
|
 |
100.0 |
26 |
PDB header:transferase
Chain: A: PDB Molecule:2-c-methyl-d-erythritol 4-phosphate cytidylyltransferase;
PDBTitle: crystal structure of francisella tularensis 2-c-methyl-d-erythritol 4-2 phosphate cytidylyltransferase (ispd)
|
|
|
|
| 6 | c4kt7A_
|
|
 |
100.0 |
26 |
PDB header:transferase
Chain: A: PDB Molecule:2-c-methyl-d-erythritol 4-phosphate cytidylyltransferase;
PDBTitle: the crystal structure of 4-diphosphocytidyl-2c-methyl-d-2 erythritolsynthase from anaerococcus prevotii dsm 20548
|
|
|
|
| 7 | c3okrA_
|
|
 |
100.0 |
89 |
PDB header:transferase
Chain: A: PDB Molecule:2-c-methyl-d-erythritol 4-phosphate cytidylyltransferase;
PDBTitle: structure of mtb apo 2-c-methyl-d-erythritol 4-phosphate2 cytidyltransferase (ispd)
|
|
|
|
| 8 | c1w57A_
|
|
 |
100.0 |
29 |
PDB header:transferase
Chain: A: PDB Molecule:ispd/ispf bifunctional enzyme;
PDBTitle: structure of the bifunctional ispdf from campylobacter2 jejuni containing zn
|
|
|
|
| 9 | d1w55a1
|
|
 |
100.0 |
28 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:Cytidylytransferase |
|
|
|
| 10 | d2oi6a2
|
|
 |
100.0 |
18 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:UDP-glucose pyrophosphorylase |
|
|
|
| 11 | c3f1cB_
|
|
 |
100.0 |
25 |
PDB header:transferase
Chain: B: PDB Molecule:putative 2-c-methyl-d-erythritol 4-phosphate
PDBTitle: crystal structure of 2-c-methyl-d-erythritol 4-phosphate2 cytidylyltransferase from listeria monocytogenes
|
|
|
|
| 12 | c4cvhA_
|
|
 |
100.0 |
28 |
PDB header:transferase
Chain: A: PDB Molecule:isoprenoid synthase domain-containing protein;
PDBTitle: crystal structure of human isoprenoid synthase domain-containing2 protein
|
|
|
|
| 13 | c3okrC_
|
|
 |
100.0 |
97 |
PDB header:transferase
Chain: C: PDB Molecule:2-c-methyl-d-erythritol 4-phosphate cytidylyltransferase;
PDBTitle: structure of mtb apo 2-c-methyl-d-erythritol 4-phosphate2 cytidyltransferase (ispd)
|
|
|
|
| 14 | d1vgwa_
|
|
 |
100.0 |
33 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:Cytidylytransferase |
|
|
|
| 15 | c2px7A_
|
|
 |
100.0 |
37 |
PDB header:transferase
Chain: A: PDB Molecule:2-c-methyl-d-erythritol 4-phosphate cytidylyltransferase;
PDBTitle: crystal structure of 2-c-methyl-d-erythritol 4-phosphate2 cytidylyltransferase from thermus thermophilus hb8
|
|
|
|
| 16 | c4ys8B_
|
|
 |
100.0 |
29 |
PDB header:transferase
Chain: B: PDB Molecule:2-c-methyl-d-erythritol 4-phosphate cytidylyltransferase;
PDBTitle: crystal structure of 2-c-methyl-d-erythritol 4-phosphate2 cytidylyltransferase (ispd) from burkholderia thailandensis
|
|
|
|
| 17 | c4xwiA_
|
|
 |
100.0 |
24 |
PDB header:transferase
Chain: A: PDB Molecule:3-deoxy-manno-octulosonate cytidylyltransferase;
PDBTitle: x-ray crystal structure of cmp-kdo synthase from pseudomonas2 aeruginosa
|
|
|
|
| 18 | d1g97a2
|
|
 |
100.0 |
16 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:UDP-glucose pyrophosphorylase |
|
|
|
| 19 | d1w77a1
|
|
 |
100.0 |
25 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:Cytidylytransferase |
|
|
|
| 20 | c3polA_
|
|
 |
100.0 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:3-deoxy-manno-octulosonate cytidylyltransferase;
PDBTitle: 2.3 angstrom crystal structure of 3-deoxy-manno-octulosonate2 cytidylyltransferase (kdsb) from acinetobacter baumannii.
|
|
|
|
| 21 | c3oamD_ |
|
not modelled |
100.0 |
18 |
PDB header:transferase
Chain: D: PDB Molecule:3-deoxy-manno-octulosonate cytidylyltransferase;
PDBTitle: crystal structure of cytidylyltransferase from vibrio cholerae
|
|
|
| 22 | d1vica_ |
|
not modelled |
100.0 |
18 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:Cytidylytransferase |
|
|
| 23 | c2vshB_ |
|
not modelled |
100.0 |
25 |
PDB header:transferase
Chain: B: PDB Molecule:2-c-methyl-d-erythritol 4-phosphate
PDBTitle: synthesis of cdp-activated ribitol for teichoic acid2 precursors in streptococcus pneumoniae
|
|
|
| 24 | d1h7ea_ |
|
not modelled |
100.0 |
19 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:Cytidylytransferase |
|
|
| 25 | c3d8vA_ |
|
not modelled |
100.0 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:bifunctional protein glmu;
PDBTitle: crystal structure of glmu from mycobacterium tuberculosis2 in complex with uridine-diphosphate-n-acetylglucosamine
|
|
|
| 26 | c4jisB_ |
|
not modelled |
100.0 |
26 |
PDB header:transferase
Chain: B: PDB Molecule:ribitol-5-phosphate cytidylyltransferase;
PDBTitle: crystal structure of ribitol 5-phosphate cytidylyltransferase (tari)2 from bacillus subtilis
|
|
|
| 27 | c3pnnA_ |
|
not modelled |
100.0 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:conserved domain protein;
PDBTitle: the crystal structure of a glycosyltransferase from porphyromonas2 gingivalis w83
|
|
|
| 28 | d1qwja_ |
|
not modelled |
100.0 |
15 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:Cytidylytransferase |
|
|
| 29 | c3hl3A_ |
|
not modelled |
100.0 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:glucose-1-phosphate thymidylyltransferase;
PDBTitle: 2.76 angstrom crystal structure of a putative glucose-1-phosphate2 thymidylyltransferase from bacillus anthracis in complex with a3 sucrose.
|
|
|
| 30 | d1eyra_ |
|
not modelled |
100.0 |
15 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:Cytidylytransferase |
|
|
| 31 | d1h5ra_ |
|
not modelled |
100.0 |
15 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:glucose-1-phosphate thymidylyltransferase |
|
|
| 32 | c2wawA_ |
|
not modelled |
99.9 |
17 |
PDB header:unknown function
Chain: A: PDB Molecule:moba relate protein;
PDBTitle: crystal structure of mycobacterium tuberculosis rv0371c2 homolog from mycobacterium sp. strain jc1
|
|
|
| 33 | d1fxoa_ |
|
not modelled |
99.9 |
13 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:glucose-1-phosphate thymidylyltransferase |
|
|
| 34 | d1iina_ |
|
not modelled |
99.9 |
14 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:glucose-1-phosphate thymidylyltransferase |
|
|
| 35 | c2y6pC_ |
|
not modelled |
99.9 |
15 |
PDB header:transferase
Chain: C: PDB Molecule:3-deoxy-manno-octulosonate cytidylyltransferase;
PDBTitle: evidence for a two-metal-ion-mechanism in the kdo-2 cytidylyltransferase kdsb
|
|
|
| 36 | c3tqdA_ |
|
not modelled |
99.9 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:3-deoxy-manno-octulosonate cytidylyltransferase;
PDBTitle: structure of the 3-deoxy-d-manno-octulosonate cytidylyltransferase2 (kdsb) from coxiella burnetii
|
|
|
| 37 | c6oewB_ |
|
not modelled |
99.9 |
11 |
PDB header:transferase
Chain: B: PDB Molecule:cytidylyltransferase;
PDBTitle: structure of a cytidylyltransferase from leptospira borgpetersenii2 serovar hardjo-bovis (strain jb197)
|
|
|
| 38 | c4jd0A_ |
|
not modelled |
99.9 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:nucleotidyl transferase;
PDBTitle: structure of the inositol-1-phosphate ctp transferase from t.2 maritima.
|
|
|
| 39 | c2we9A_ |
|
not modelled |
99.9 |
14 |
PDB header:unknown function
Chain: A: PDB Molecule:moba-related protein;
PDBTitle: crystal structure of rv0371c from mycobacterium2 tuberculosis h37rv
|
|
|
| 40 | d1mc3a_ |
|
not modelled |
99.9 |
14 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:glucose-1-phosphate thymidylyltransferase |
|
|
| 41 | c5i1fA_ |
|
not modelled |
99.9 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:utp--glucose-1-phosphate uridylyltransferase;
PDBTitle: crystal structure of utp-glucose-1-phosphate uridylyltransferase from2 burkholderia vietnamiensis in complex with uridine-5'-diphosphate-3 glucose
|
|
|
| 42 | c5xhwA_ |
|
not modelled |
99.9 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:putative 6-deoxy-d-mannoheptose pathway protein;
PDBTitle: crystal structure of hddc from yersinia pseudotuberculosis
|
|
|
| 43 | c2pa4B_ |
|
not modelled |
99.9 |
16 |
PDB header:transferase
Chain: B: PDB Molecule:utp-glucose-1-phosphate uridylyltransferase;
PDBTitle: crystal structure of udp-glucose pyrophosphorylase from corynebacteria2 glutamicum in complex with magnesium and udp-glucose
|
|
|
| 44 | d1lvwa_ |
|
not modelled |
99.9 |
16 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:glucose-1-phosphate thymidylyltransferase |
|
|
| 45 | d1vh1a_ |
|
not modelled |
99.9 |
17 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:Cytidylytransferase |
|
|
| 46 | c3foqA_ |
|
not modelled |
99.9 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:bifunctional protein glmu;
PDBTitle: crystal structure of n-acetylglucosamine-1-phosphate2 uridyltransferase (glmu) from mycobacterium tuberculosis in3 a cubic space group.
|
|
|
| 47 | d1e5ka_ |
|
not modelled |
99.9 |
18 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:Molybdenum cofactor biosynthesis protein MobA |
|
|
| 48 | c3jukA_ |
|
not modelled |
99.9 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:udp-glucose pyrophosphorylase (galu);
PDBTitle: the crystal structure of udp-glucose pyrophosphorylase complexed with2 udp-glucose
|
|
|
| 49 | c4y7uA_ |
|
not modelled |
99.9 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:nucleotidyl transferase;
PDBTitle: structural analysis of muru
|
|
|
| 50 | c2oi6A_ |
|
not modelled |
99.9 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:bifunctional protein glmu;
PDBTitle: e. coli glmu- complex with udp-glcnac, coa and glcn-1-po4
|
|
|
| 51 | c2v0hA_ |
|
not modelled |
99.9 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:bifunctional protein glmu;
PDBTitle: characterization of substrate binding and catalysis of the2 potential antibacterial target n-acetylglucosamine-1-3 phosphate uridyltransferase (glmu)
|
|
|
| 52 | c1hm8A_ |
|
not modelled |
99.9 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:udp-n-acetylglucosamine-1-phosphate uridyltransferase;
PDBTitle: crystal structure of s.pneumoniae n-acetylglucosamine-1-phosphate2 uridyltransferase, glmu, bound to acetyl coenzyme a
|
|
|
| 53 | c4mndA_ |
|
not modelled |
99.9 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:ctp l-myo-inositol-1-phosphate cytidylyltransferase/cdp-l-
PDBTitle: crystal structure of archaeoglobus fulgidus ipct-dipps bifunctional2 membrane protein
|
|
|
| 54 | c3d98A_ |
|
not modelled |
99.9 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:bifunctional protein glmu;
PDBTitle: crystal structure of glmu from mycobacterium tuberculosis, ligand-free2 form
|
|
|
| 55 | c2ux8G_ |
|
not modelled |
99.9 |
17 |
PDB header:transferase
Chain: G: PDB Molecule:glucose-1-phosphate uridylyltransferase;
PDBTitle: crystal structure of sphingomonas elodea atcc 31461 glucose-2 1-phosphate uridylyltransferase in complex with glucose-3 1-phosphate.
|
|
|
| 56 | c1jylC_ |
|
not modelled |
99.9 |
18 |
PDB header:transferase
Chain: C: PDB Molecule:ctp:phosphocholine cytidylytransferase;
PDBTitle: catalytic mechanism of ctp:phosphocholine2 cytidylytransferase from streptococcus pneumoniae (licc)
|
|
|
| 57 | d1jyka_ |
|
not modelled |
99.9 |
18 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:Cytidylytransferase |
|
|
| 58 | c4evwB_ |
|
not modelled |
99.9 |
14 |
PDB header:transferase
Chain: B: PDB Molecule:nucleoside-diphosphate-sugar pyrophosphorylase;
PDBTitle: crystal structure of the nucleoside-diphosphate-sugar2 pyrophosphorylase from vibrio cholerae rc9. northeast structural3 genomics consortium (nesg) target vcr193.
|
|
|
| 59 | d2dpwa1 |
|
not modelled |
99.9 |
16 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:TTHA0179-like |
|
|
| 60 | c6b5kA_ |
|
not modelled |
99.9 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:glucose-1-phosphate thymidylyltransferase;
PDBTitle: mycobacterium tuberculosis rmla in complex with mg/dttp
|
|
|
| 61 | c1fwyA_ |
|
not modelled |
99.9 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:udp-n-acetylglucosamine pyrophosphorylase;
PDBTitle: crystal structure of n-acetylglucosamine 1-phosphate2 uridyltransferase bound to udp-glcnac
|
|
|
| 62 | d2cu2a2 |
|
not modelled |
99.9 |
21 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:mannose-1-phosphate guanylyl transferase |
|
|
| 63 | d1tzfa_ |
|
not modelled |
99.9 |
10 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:Cytidylytransferase |
|
|
| 64 | c2qkxA_ |
|
not modelled |
99.9 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:bifunctional protein glmu;
PDBTitle: n-acetyl glucosamine 1-phosphate uridyltransferase from mycobacterium2 tuberculosis complex with n-acetyl glucosamine 1-phosphate
|
|
|
| 65 | c3ngwA_ |
|
not modelled |
99.9 |
15 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:molybdopterin-guanine dinucleotide biosynthesis protein a
PDBTitle: crystal structure of molybdopterin-guanine dinucleotide biosynthesis2 protein a from archaeoglobus fulgidus, northeast structural genomics3 consortium target gr189
|
|
|
| 66 | c2e3dB_ |
|
not modelled |
99.9 |
18 |
PDB header:transferase
Chain: B: PDB Molecule:utp--glucose-1-phosphate uridylyltransferase;
PDBTitle: crystal structure of e. coli glucose-1-phosphate2 uridylyltransferase
|
|
|
| 67 | c1yp3C_ |
|
not modelled |
99.9 |
17 |
PDB header:transferase
Chain: C: PDB Molecule:glucose-1-phosphate adenylyltransferase small
PDBTitle: crystal structure of potato tuber adp-glucose2 pyrophosphorylase in complex with atp
|
|
|
| 68 | c6ifdD_ |
|
not modelled |
99.9 |
17 |
PDB header:sugar binding protein
Chain: D: PDB Molecule:cmp-n-acetylneuraminate synthetase;
PDBTitle: crystal structure of cmp-n-acetylneuraminate synthetase from vibrio2 cholerae in complex with cdp and mg2+.
|
|
|
| 69 | c3rsbB_ |
|
not modelled |
99.9 |
22 |
PDB header:transferase
Chain: B: PDB Molecule:adenosylcobinamide-phosphate guanylyltransferase;
PDBTitle: structure of the archaeal gtp:adocbi-p guanylyltransferase (coby) from2 methanocaldococcus jannaschii
|
|
|
| 70 | c2x5sB_ |
|
not modelled |
99.9 |
18 |
PDB header:transferase
Chain: B: PDB Molecule:mannose-1-phosphate guanylyltransferase;
PDBTitle: crystal structure of t. maritima gdp-mannose2 pyrophosphorylase in apo state.
|
|
|
| 71 | d1yp2a2 |
|
not modelled |
99.9 |
17 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:glucose-1-phosphate thymidylyltransferase |
|
|
| 72 | c2cu2A_ |
|
not modelled |
99.9 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:putative mannose-1-phosphate guanylyl transferase;
PDBTitle: crystal structure of mannose-1-phosphate geranyltransferase from2 thermus thermophilus hb8
|
|
|
| 73 | c5vmkB_ |
|
not modelled |
99.9 |
14 |
PDB header:transferase
Chain: B: PDB Molecule:bifunctional protein glmu;
PDBTitle: crystal structure of a bifunctional glmu udp-n-acetylglucosamine2 diphosphorylase/glucosamine-1- phosphate n-acetyltransferase from3 acinetobacter baumannii
|
|
|
| 74 | c5l6sF_ |
|
not modelled |
99.9 |
14 |
PDB header:transferase
Chain: F: PDB Molecule:glucose-1-phosphate adenylyltransferase;
PDBTitle: crystal structure of e. coli adp-glucose pyrophosphorylase (agpase) in2 complex with a positive allosteric regulator beta-fructose-1,6-3 diphosphate (fbp) - agpase*fbp
|
|
|
| 75 | c3brkX_ |
|
not modelled |
99.8 |
15 |
PDB header:transferase
Chain: X: PDB Molecule:glucose-1-phosphate adenylyltransferase;
PDBTitle: crystal structure of adp-glucose pyrophosphorylase from agrobacterium2 tumefaciens
|
|
|
| 76 | c6i3mG_ |
|
not modelled |
99.8 |
10 |
PDB header:translation
Chain: G: PDB Molecule:translation initiation factor eif-2b subunit epsilon;
PDBTitle: eif2b:eif2 complex, phosphorylated on eif2 alpha serine 52.
|
|
|
| 77 | c2ggqA_ |
|
not modelled |
99.8 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:401aa long hypothetical glucose-1-phosphate
PDBTitle: complex of hypothetical glucose-1-phosphate thymidylyltransferase from2 sulfolobus tokodaii
|
|
|
| 78 | c5b04I_ |
|
not modelled |
99.8 |
11 |
PDB header:translation
Chain: I: PDB Molecule:probable translation initiation factor eif-2b subunit
PDBTitle: crystal structure of the eukaryotic translation initiation factor 2b2 from schizosaccharomyces pombe
|
|
|
| 79 | c2e8bA_ |
|
not modelled |
99.7 |
19 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:probable molybdopterin-guanine dinucleotide biosynthesis
PDBTitle: crystal structure of the putative protein (aq1419) from aquifex2 aeolicus vf5
|
|
|
| 80 | c2qh5B_ |
|
not modelled |
99.7 |
12 |
PDB header:isomerase
Chain: B: PDB Molecule:mannose-6-phosphate isomerase;
PDBTitle: crystal structure of mannose-6-phosphate isomerase from helicobacter2 pylori
|
|
|
| 81 | c6jlwJ_ |
|
not modelled |
99.7 |
14 |
PDB header:translation
Chain: J: PDB Molecule:translation initiation factor eif-2b subunit epsilon;
PDBTitle: eif2 - eif2b complex
|
|
|
| 82 | c5b04F_ |
|
not modelled |
99.6 |
11 |
PDB header:translation
Chain: F: PDB Molecule:probable translation initiation factor eif-2b subunit
PDBTitle: crystal structure of the eukaryotic translation initiation factor 2b2 from schizosaccharomyces pombe
|
|
|
| 83 | d1vh3a_ |
|
not modelled |
99.6 |
14 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:Cytidylytransferase |
|
|
| 84 | c6bwhB_ |
|
not modelled |
99.6 |
17 |
PDB header:transferase
Chain: B: PDB Molecule:2-phospho-l-lactate guanylyltransferase;
PDBTitle: crystal structure of mycoibacterium tuberculosis rv2983 in complex2 with pep
|
|
|
| 85 | c6ezoF_ |
|
not modelled |
99.6 |
18 |
PDB header:membrane protein
Chain: F: PDB Molecule:translation initiation factor eif-2b subunit gamma;
PDBTitle: eukaryotic initiation factor eif2b in complex with isrib
|
|
|
| 86 | c2xmhB_ |
|
not modelled |
99.6 |
12 |
PDB header:transferase
Chain: B: PDB Molecule:ctp-inositol-1-phosphate cytidylyltransferase;
PDBTitle: the x-ray structure of ctp:inositol-1-phosphate cytidylyltransferase2 from archaeoglobus fulgidus
|
|
|
| 87 | c3d5nB_ |
|
not modelled |
99.6 |
16 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:q97w15_sulso;
PDBTitle: crystal structure of the q97w15_sulso protein from sulfolobus2 solfataricus. nesg target ssr125.
|
|
|
| 88 | c6qg2F_ |
|
not modelled |
99.5 |
14 |
PDB header:translation
Chain: F: PDB Molecule:translation initiation factor eif-2b subunit gamma;
PDBTitle: structure of eif2b-eif2 (phosphorylated at ser51) complex (model a)
|
|
|
| 89 | c6ezoJ_ |
|
not modelled |
99.0 |
14 |
PDB header:membrane protein
Chain: J: PDB Molecule:human eukaryotic initiation factor eif2b epsilon subunits;
PDBTitle: eukaryotic initiation factor eif2b in complex with isrib
|
|
|
| 90 | d2icya2 |
|
not modelled |
98.6 |
14 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:UDP-glucose pyrophosphorylase |
|
|
| 91 | c2q4jB_ |
|
not modelled |
98.6 |
14 |
PDB header:transferase
Chain: B: PDB Molecule:probable utp-glucose-1-phosphate uridylyltransferase 2;
PDBTitle: ensemble refinement of the protein crystal structure of gene product2 from arabidopsis thaliana at3g03250, a putative udp-glucose3 pyrophosphorylase
|
|
|
| 92 | c3oc9A_ |
|
not modelled |
98.5 |
31 |
PDB header:transferase
Chain: A: PDB Molecule:udp-n-acetylglucosamine pyrophosphorylase;
PDBTitle: crystal structure of putative udp-n-acetylglucosamine2 pyrophosphorylase from entamoeba histolytica
|
|
|
| 93 | c3r2wB_ |
|
not modelled |
98.5 |
12 |
PDB header:transferase
Chain: B: PDB Molecule:utp--glucose-1-phosphate uridylyltransferase;
PDBTitle: crystal strucutre of udp-glucose pyrophosphorylase of homo sapiens
|
|
|
| 94 | c3cgxA_ |
|
not modelled |
98.5 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:putative nucleotide-diphospho-sugar transferase;
PDBTitle: crystal structure of putative nucleotide-diphospho-sugar transferase2 (yp_389115.1) from desulfovibrio desulfuricans g20 at 1.90 a3 resolution
|
|
|
| 95 | c6i7tI_ |
|
not modelled |
98.5 |
9 |
PDB header:translation
Chain: I: PDB Molecule:translation initiation factor eif-2b subunit gamma;
PDBTitle: eif2b:eif2 complex
|
|
|
| 96 | c2i5kB_ |
|
not modelled |
98.4 |
12 |
PDB header:transferase
Chain: B: PDB Molecule:utp--glucose-1-phosphate uridylyltransferase;
PDBTitle: crystal structure of ugp1p
|
|
|
| 97 | c4bmaB_ |
|
not modelled |
98.4 |
21 |
PDB header:transferase
Chain: B: PDB Molecule:udp-n-acetylglucosamine pyrophosphorylase;
PDBTitle: structural of aspergillus fumigatus udp-n-acetylglucosamine2 pyrophosphorylase
|
|
|
| 98 | c3ogzA_ |
|
not modelled |
98.3 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:udp-sugar pyrophosphorylase;
PDBTitle: protein structure of usp from l. major in apo-form
|
|
|
| 99 | c2yqsA_ |
|
not modelled |
98.3 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:udp-n-acetylglucosamine pyrophosphorylase;
PDBTitle: crystal structure of uridine-diphospho-n-acetylglucosamine2 pyrophosphorylase from candida albicans, in the product-binding form
|
|
|
| 100 | d1jv1a_ |
|
not modelled |
98.3 |
31 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:UDP-glucose pyrophosphorylase |
|
|
| 101 | d1vm8a_ |
|
not modelled |
98.3 |
30 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:UDP-glucose pyrophosphorylase |
|
|
| 102 | c2oefA_ |
|
not modelled |
98.2 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:utp-glucose-1-phosphate uridylyltransferase 2,
PDBTitle: open and closed structures of the udp-glucose2 pyrophosphorylase from leishmania major
|
|
|
| 103 | c3gueB_ |
|
not modelled |
98.2 |
24 |
PDB header:transferase
Chain: B: PDB Molecule:utp-glucose-1-phosphate uridylyltransferase 2;
PDBTitle: crystal structure of udp-glucose phosphorylase from trypanosoma2 brucei, (tb10.389.0330)
|
|
|
| 104 | d2i5ea1 |
|
not modelled |
98.1 |
16 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:MM2497-like |
|
|
| 105 | c4bqhA_ |
|
not modelled |
98.1 |
29 |
PDB header:transferase
Chain: A: PDB Molecule:udp-n-acetylglucosamine pyrophosphorylase;
PDBTitle: crystal structure of the uridine diphosphate n-2 acetylglucosamine pyrophosphorylase from trypanosoma3 brucei in complex with inhibitor
|
|
|
| 106 | c5tz8C_ |
|
not modelled |
96.3 |
12 |
PDB header:transferase
Chain: C: PDB Molecule:glycosyl transferase;
PDBTitle: crystal structure of s. aureus tars
|
|
|
| 107 | c5mm1A_ |
|
not modelled |
95.6 |
14 |
PDB header:membrane protein
Chain: A: PDB Molecule:dolichol monophosphate mannose synthase;
PDBTitle: dolichyl phosphate mannose synthase in complex with gdp and dolichyl2 phosphate mannose
|
|
|
| 108 | c5ekeB_ |
|
not modelled |
95.1 |
19 |
PDB header:transferase
Chain: B: PDB Molecule:uncharacterized glycosyltransferase sll0501;
PDBTitle: structure of the polyisoprenyl-phosphate glycosyltransferase gtrb2 (f215a mutant)
|
|
|
| 109 | d1omza_ |
|
not modelled |
94.6 |
12 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:Exostosin |
|
|
| 110 | c2ffuA_ |
|
not modelled |
94.3 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:polypeptide n-acetylgalactosaminyltransferase 2;
PDBTitle: crystal structure of human ppgalnact-2 complexed with udp and ea2
|
|
|
| 111 | c6h4mA_ |
|
not modelled |
94.3 |
7 |
PDB header:transferase
Chain: A: PDB Molecule:probable ss-1,3-n-acetylglucosaminyltransferase;
PDBTitle: tarp-udp-glcnac-3rbop
|
|
|
| 112 | c2z86D_ |
|
not modelled |
94.2 |
12 |
PDB header:transferase
Chain: D: PDB Molecule:chondroitin synthase;
PDBTitle: crystal structure of chondroitin polymerase from escherichia coli2 strain k4 (k4cp) complexed with udp-glcua and udp
|
|
|
| 113 | d1xhba2 |
|
not modelled |
94.0 |
9 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:Polypeptide N-acetylgalactosaminyltransferase 1, N-terminal domain |
|
|
| 114 | d2bo4a1 |
|
not modelled |
93.3 |
16 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:MGS-like |
|
|
| 115 | c2d7iA_ |
|
not modelled |
92.7 |
8 |
PDB header:transferase
Chain: A: PDB Molecule:polypeptide n-acetylgalactosaminyltransferase 10;
PDBTitle: crystal structure of pp-galnac-t10 with udp, galnac and mn2+
|
|
|
| 116 | c4hg6A_ |
|
not modelled |
92.5 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:cellulose synthase subunit a;
PDBTitle: structure of a cellulose synthase - cellulose translocation2 intermediate
|
|
|
| 117 | c1omxB_ |
|
not modelled |
92.3 |
10 |
PDB header:transferase
Chain: B: PDB Molecule:alpha-1,4-n-acetylhexosaminyltransferase extl2;
PDBTitle: crystal structure of mouse alpha-1,4-n-acetylhexosaminyltransferase2 (extl2)
|
|
|
| 118 | c3ckvA_ |
|
not modelled |
90.9 |
17 |
PDB header:unknown function
Chain: A: PDB Molecule:putative uncharacterized protein;
PDBTitle: crystal structure of a mycobacterial protein
|
|
|
| 119 | c6e4rB_ |
|
not modelled |
90.8 |
9 |
PDB header:transferase
Chain: B: PDB Molecule:polypeptide n-acetylgalactosaminyltransferase 9;
PDBTitle: crystal structure of the drosophila melanogaster polypeptide n-2 acetylgalactosaminyl transferase pgant9b
|
|
|
| 120 | c3f1yC_ |
|
not modelled |
90.7 |
17 |
PDB header:transferase
Chain: C: PDB Molecule:mannosyl-3-phosphoglycerate synthase;
PDBTitle: mannosyl-3-phosphoglycerate synthase from rubrobacter xylanophilus
|
|
|