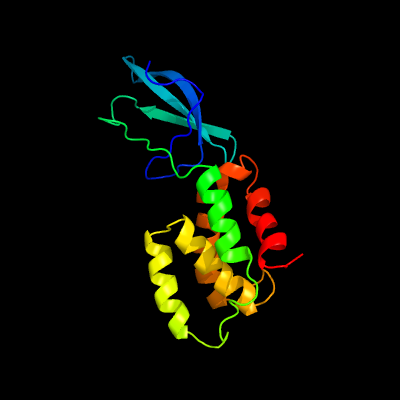
| 1 | c2lwjA_
|
|
 |
100.0 |
32 |
PDB header:transcription
Chain: A: PDB Molecule:transcriptional regulator, card family;
PDBTitle: nmr solution structure myxoccoccus xanthus cdnl
|
|
|
|
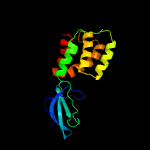
| 2 | c4l5gA_
|
|
 |
100.0 |
25 |
PDB header:transcription
Chain: A: PDB Molecule:card;
PDBTitle: crystal structure of thermus thermophilus card
|
|
|
|
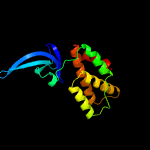
| 3 | c4kbmB_
|
|
 |
100.0 |
99 |
PDB header:transferase/transcription
Chain: B: PDB Molecule:rna polymerase-binding transcription factor card;
PDBTitle: structure of the mtb card/rnap beta subunit b1-b2 domains complex
|
|
|
|
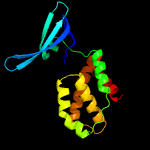
| 4 | c2lt3A_
|
|
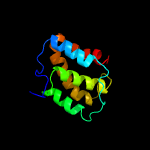 |
99.9 |
33 |
PDB header:transcription
Chain: A: PDB Molecule:transcriptional regulator, card family;
PDBTitle: solution nmr structure of the c-terminal domain of cdnl from2 myxococcus xanthus
|
|
|
|
| 5 | c2eyqA_
|
|
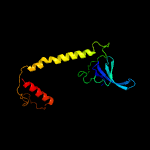 |
99.9 |
21 |
PDB header:hydrolase
Chain: A: PDB Molecule:transcription-repair coupling factor;
PDBTitle: crystal structure of escherichia coli transcription-repair2 coupling factor
|
|
|
|
| 6 | c2lt4A_
|
|
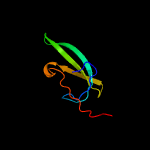 |
99.8 |
29 |
PDB header:transcription
Chain: A: PDB Molecule:transcriptional regulator, card family;
PDBTitle: cdnlnt from myxoccoccus xanthus
|
|
|
|
| 7 | c2lt1A_
|
|
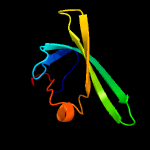 |
99.8 |
31 |
PDB header:transcription
Chain: A: PDB Molecule:card protein;
PDBTitle: solution nmr structure of the 72-residue n-terminal domain of2 myxococcus xanthus card
|
|
|
|
| 8 | c2lqkA_
|
|
 |
99.8 |
29 |
PDB header:transcription regulator
Chain: A: PDB Molecule:transcriptional regulator;
PDBTitle: nmr solution structure of the n-terminal domain of the cdnl protein2 from thermus thermophilus
|
|
|
|
| 9 | d2eyqa1
|
|
 |
99.6 |
23 |
Fold:SH3-like barrel
Superfamily:CarD-like
Family:CarD-like |
|
|
|
| 10 | c3mlqE_
|
|
 |
98.1 |
27 |
PDB header:transferase/transcription
Chain: E: PDB Molecule:transcription-repair coupling factor;
PDBTitle: crystal structure of the thermus thermophilus transcription-repair2 coupling factor rna polymerase interacting domain with the thermus3 aquaticus rna polymerase beta1 domain
|
|
|
|
| 11 | c3mlqH_
|
|
 |
98.0 |
24 |
PDB header:transferase/transcription
Chain: H: PDB Molecule:transcription-repair coupling factor;
PDBTitle: crystal structure of the thermus thermophilus transcription-repair2 coupling factor rna polymerase interacting domain with the thermus3 aquaticus rna polymerase beta1 domain
|
|
|
|
| 12 | c5dmaA_
|
|
 |
94.2 |
29 |
PDB header:hydrolase
Chain: A: PDB Molecule:atp-dependent dna helicase pcra;
PDBTitle: crystal structure of c-terminal tudor domain in pcra/uvrd helicase
|
|
|
|
| 13 | c3t9nG_
|
|
 |
70.0 |
24 |
PDB header:membrane protein
Chain: G: PDB Molecule:small-conductance mechanosensitive channel;
PDBTitle: crystal structure of a membrane protein
|
|
|
|
| 14 | d2vv5a1
|
|
 |
52.5 |
24 |
Fold:Sm-like fold
Superfamily:Sm-like ribonucleoproteins
Family:Mechanosensitive channel protein MscS (YggB), middle domain |
|
|
|
| 15 | d2eyqa3
|
|
 |
52.1 |
17 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Tandem AAA-ATPase domain |
|
|
|
| 16 | d1in4a1
|
|
 |
49.5 |
20 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:Helicase DNA-binding domain |
|
|
|
| 17 | c2lrqA_
|
|
 |
49.5 |
22 |
PDB header:transcription
Chain: A: PDB Molecule:nua4 complex subunit eaf3 homolog;
PDBTitle: chemical shift assignment and solution structure of fr822a from2 drosophila melanogaster. northeast structural genomics consortium3 target fr822a
|
|
|
|
| 18 | c6au8A_
|
|
 |
48.9 |
16 |
PDB header:chaperone
Chain: A: PDB Molecule:golgi to er traffic protein 4 homolog;
PDBTitle: 1.8 angstrom crystal structure of the human bag6-nls & trc35 complex
|
|
|
|
| 19 | c2vv5D_
|
|
 |
44.0 |
24 |
PDB header:membrane protein
Chain: D: PDB Molecule:small-conductance mechanosensitive channel;
PDBTitle: the open structure of mscs
|
|
|
|
| 20 | c4hw9E_
|
|
 |
42.7 |
27 |
PDB header:membrane protein
Chain: E: PDB Molecule:mechanosensitive channel mscs;
PDBTitle: crystal structure of helicobacter pylori mscs (closed state)
|
|
|
|
| 21 | c6ghbB_ |
|
not modelled |
40.1 |
14 |
PDB header:protein binding
Chain: B: PDB Molecule:upf0413 protein gk0824;
PDBTitle: crystal structure of spx in complex with yjbh (oxidized)
|
|
|
| 22 | c5y4oA_ |
|
not modelled |
40.1 |
27 |
PDB header:membrane protein
Chain: A: PDB Molecule:low conductance mechanosensitive channel ynai;
PDBTitle: cryo-em structure of mscs channel, ynai
|
|
|
| 23 | d1aono_ |
|
not modelled |
36.4 |
29 |
Fold:GroES-like
Superfamily:GroES-like
Family:GroES |
|
|
| 24 | c3nx6A_ |
|
not modelled |
35.7 |
37 |
PDB header:chaperone
Chain: A: PDB Molecule:10kda chaperonin;
PDBTitle: crystal structure of co-chaperonin, groes (xoo4289) from xanthomonas2 oryzae pv. oryzae kacc10331
|
|
|
| 25 | d1vbva1 |
|
not modelled |
34.9 |
14 |
Fold:SH3-like barrel
Superfamily:YccV-like
Family:YccV-like |
|
|
| 26 | d1p3ha_ |
|
not modelled |
31.0 |
30 |
Fold:GroES-like
Superfamily:GroES-like
Family:GroES |
|
|
| 27 | d1lepa_ |
|
not modelled |
29.2 |
33 |
Fold:GroES-like
Superfamily:GroES-like
Family:GroES |
|
|
| 28 | d1ixrc1 |
|
not modelled |
29.1 |
28 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:Helicase DNA-binding domain |
|
|
| 29 | c5in1A_ |
|
not modelled |
27.9 |
24 |
PDB header:transcription
Chain: A: PDB Molecule:mrg701;
PDBTitle: crystal structure of the mrg701 chromodomain
|
|
|
| 30 | c1w4sA_ |
|
not modelled |
27.1 |
24 |
PDB header:nuclear protein
Chain: A: PDB Molecule:polybromo 1 protein;
PDBTitle: crystal structure of the proximal bah domain of polybromo
|
|
|
| 31 | c4pl6A_ |
|
not modelled |
26.1 |
21 |
PDB header:transcription
Chain: A: PDB Molecule:at1g02740;
PDBTitle: structure of the chromodomain of mrg2 in complex with h3k4me3
|
|
|
| 32 | c2wpvG_ |
|
not modelled |
25.5 |
15 |
PDB header:protein binding
Chain: G: PDB Molecule:upf0363 protein yor164c;
PDBTitle: crystal structure of s. cerevisiae get4-get5 complex
|
|
|
| 33 | d1j20a2 |
|
not modelled |
25.3 |
23 |
Fold:Argininosuccinate synthetase, C-terminal domain
Superfamily:Argininosuccinate synthetase, C-terminal domain
Family:Argininosuccinate synthetase, C-terminal domain |
|
|
| 34 | d1vl2a2 |
|
not modelled |
24.1 |
18 |
Fold:Argininosuccinate synthetase, C-terminal domain
Superfamily:Argininosuccinate synthetase, C-terminal domain
Family:Argininosuccinate synthetase, C-terminal domain |
|
|
| 35 | d1we3o_ |
|
not modelled |
22.8 |
35 |
Fold:GroES-like
Superfamily:GroES-like
Family:GroES |
|
|
| 36 | c4pj1V_ |
|
not modelled |
22.3 |
33 |
PDB header:chaperone
Chain: V: PDB Molecule:10 kda heat shock protein, mitochondrial;
PDBTitle: crystal structure of the human mitochondrial chaperonin symmetrical2 'football' complex
|
|
|
| 37 | d1k92a2 |
|
not modelled |
21.6 |
25 |
Fold:Argininosuccinate synthetase, C-terminal domain
Superfamily:Argininosuccinate synthetase, C-terminal domain
Family:Argininosuccinate synthetase, C-terminal domain |
|
|
| 38 | d2i5nh1 |
|
not modelled |
21.6 |
18 |
Fold:PRC-barrel domain
Superfamily:PRC-barrel domain
Family:Photosynthetic reaction centre, H-chain, cytoplasmic domain |
|
|
| 39 | d1iz6a1 |
|
not modelled |
21.4 |
16 |
Fold:SH3-like barrel
Superfamily:Translation proteins SH3-like domain
Family:eIF5a N-terminal domain-like |
|
|
| 40 | c5xonW_ |
|
not modelled |
21.1 |
17 |
PDB header:transcription/rna
Chain: W: PDB Molecule:protein that forms a complex with spt4p;
PDBTitle: rna polymerase ii elongation complex bound with spt4/5 and tfiis
|
|
|
| 41 | c3e20C_ |
|
not modelled |
20.9 |
8 |
PDB header:translation
Chain: C: PDB Molecule:eukaryotic peptide chain release factor subunit 1;
PDBTitle: crystal structure of s.pombe erf1/erf3 complex
|
|
|
| 42 | d2f5ka1 |
|
not modelled |
20.0 |
24 |
Fold:SH3-like barrel
Superfamily:Chromo domain-like
Family:Chromo barrel domain |
|
|
| 43 | c3pnwX_ |
|
not modelled |
18.6 |
15 |
PDB header:protein binding/immune system
Chain: X: PDB Molecule:tudor domain-containing protein 3;
PDBTitle: crystal structure of the tudor domain of human tdrd3 in complex with2 an anti-tdrd3 fab
|
|
|
| 44 | d2eifa1 |
|
not modelled |
18.2 |
20 |
Fold:SH3-like barrel
Superfamily:Translation proteins SH3-like domain
Family:eIF5a N-terminal domain-like |
|
|
| 45 | c4ytlB_ |
|
not modelled |
17.7 |
20 |
PDB header:transcription
Chain: B: PDB Molecule:transcription elongation factor spt5;
PDBTitle: structure of the kow2-kow3 domain of transcription elongation factor2 spt5.
|
|
|
| 46 | c3qtmB_ |
|
not modelled |
17.6 |
19 |
PDB header:translation
Chain: B: PDB Molecule:uncharacterized protein c4b3.07;
PDBTitle: structure of s. pombe nuclear import adaptor nro1 (space group p21)
|
|
|
| 47 | d2cwya1 |
|
not modelled |
17.1 |
22 |
Fold:Hyaluronidase domain-like
Superfamily:TTHA0068-like
Family:TTHA0068-like |
|
|
| 48 | c2ymyB_ |
|
not modelled |
16.7 |
17 |
PDB header:apoptosis
Chain: B: PDB Molecule:ras association domain-containing protein 5;
PDBTitle: structure of the murine nore1-sarah domain
|
|
|
| 49 | c2lclA_ |
|
not modelled |
16.4 |
21 |
PDB header:transcription
Chain: A: PDB Molecule:transcriptional activator rfah;
PDBTitle: solution structure of rfah carboxyterminal domain
|
|
|
| 50 | c3nojA_ |
|
not modelled |
15.2 |
32 |
PDB header:lyase
Chain: A: PDB Molecule:4-carboxy-4-hydroxy-2-oxoadipate aldolase/oxaloacetate
PDBTitle: the structure of hmg/cha aldolase from the protocatechuate degradation2 pathway of pseudomonas putida
|
|
|
| 51 | c2ko4A_ |
|
not modelled |
15.2 |
12 |
PDB header:transcription
Chain: A: PDB Molecule:mediator of rna polymerase ii transcription subunit 15;
PDBTitle: complex structure of the activation domain of gcn4 bound to the2 mediator co-activator domain of gal11/med15
|
|
|
| 52 | d1dt9a3 |
|
not modelled |
14.7 |
11 |
Fold:N-terminal domain of eukaryotic peptide chain release factor subunit 1, ERF1
Superfamily:N-terminal domain of eukaryotic peptide chain release factor subunit 1, ERF1
Family:N-terminal domain of eukaryotic peptide chain release factor subunit 1, ERF1 |
|
|
| 53 | d2ijqa1 |
|
not modelled |
14.7 |
14 |
Fold:Hyaluronidase domain-like
Superfamily:TTHA0068-like
Family:TTHA0068-like |
|
|
| 54 | d1ku3a_ |
|
not modelled |
14.4 |
19 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Sigma3 and sigma4 domains of RNA polymerase sigma factors
Family:Sigma4 domain |
|
|
| 55 | d1nppa2 |
|
not modelled |
13.8 |
13 |
Fold:SH3-like barrel
Superfamily:Translation proteins SH3-like domain
Family:N-utilization substance G protein NusG, C-terminal domain |
|
|
| 56 | c1k97A_ |
|
not modelled |
13.4 |
25 |
PDB header:ligase
Chain: A: PDB Molecule:argininosuccinate synthase;
PDBTitle: crystal structure of e. coli argininosuccinate synthetase in complex2 with aspartate and citrulline
|
|
|
| 57 | d1bkba1 |
|
not modelled |
13.3 |
25 |
Fold:SH3-like barrel
Superfamily:Translation proteins SH3-like domain
Family:eIF5a N-terminal domain-like |
|
|
| 58 | c2fg0B_ |
|
not modelled |
12.8 |
35 |
PDB header:hydrolase
Chain: B: PDB Molecule:cog0791: cell wall-associated hydrolases (invasion-
PDBTitle: crystal structure of a putative gamma-d-glutamyl-l-diamino acid2 endopeptidase (npun_r0659) from nostoc punctiforme pcc 73102 at 1.793 a resolution
|
|
|
| 59 | c3j21R_ |
|
not modelled |
12.7 |
57 |
PDB header:ribosome
Chain: R: PDB Molecule:50s ribosomal protein l21e;
PDBTitle: promiscuous behavior of proteins in archaeal ribosomes revealed by2 cryo-em: implications for evolution of eukaryotic ribosomes (50s3 ribosomal proteins)
|
|
|
| 60 | c4d10C_ |
|
not modelled |
12.6 |
8 |
PDB header:signaling protein
Chain: C: PDB Molecule:cop9 signalosome complex subunit 3;
PDBTitle: crystal structure of the cop9 signalosome
|
|
|
| 61 | d1ttya_ |
|
not modelled |
12.5 |
17 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Sigma3 and sigma4 domains of RNA polymerase sigma factors
Family:Sigma4 domain |
|
|
| 62 | d2do3a1 |
|
not modelled |
12.4 |
71 |
Fold:SH3-like barrel
Superfamily:Translation proteins SH3-like domain
Family:SPT5 KOW domain-like |
|
|
| 63 | d1ixsb1 |
|
not modelled |
12.2 |
28 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:Helicase DNA-binding domain |
|
|
| 64 | d1vqoq1 |
|
not modelled |
12.1 |
43 |
Fold:SH3-like barrel
Superfamily:Translation proteins SH3-like domain
Family:Ribosomal proteins L24p and L21e |
|
|
| 65 | c5znpB_ |
|
not modelled |
12.1 |
26 |
PDB header:gene regulation
Chain: B: PDB Molecule:short life family protein;
PDBTitle: crystal structure of ptshl in complex with an h3k4me3 peptide
|
|
|
| 66 | d1ku7a_ |
|
not modelled |
12.1 |
20 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Sigma3 and sigma4 domains of RNA polymerase sigma factors
Family:Sigma4 domain |
|
|
| 67 | d7odca1 |
|
not modelled |
11.7 |
21 |
Fold:Domain of alpha and beta subunits of F1 ATP synthase-like
Superfamily:Alanine racemase C-terminal domain-like
Family:Eukaryotic ODC-like |
|
|
| 68 | c1zljE_ |
|
not modelled |
11.5 |
17 |
PDB header:transcription
Chain: E: PDB Molecule:dormancy survival regulator;
PDBTitle: crystal structure of the mycobacterium tuberculosis hypoxic2 response regulator dosr c-terminal domain
|
|
|
| 69 | c3sztB_ |
|
not modelled |
11.3 |
26 |
PDB header:transcription
Chain: B: PDB Molecule:quorum-sensing control repressor;
PDBTitle: quorum sensing control repressor, qscr, bound to n-3-oxo-dodecanoyl-l-2 homoserine lactone
|
|
|
| 70 | c2e6zA_ |
|
not modelled |
11.1 |
57 |
PDB header:transcription
Chain: A: PDB Molecule:transcription elongation factor spt5;
PDBTitle: solution structure of the second kow motif of human2 transcription elongation factor spt5
|
|
|
| 71 | d2p7vb1 |
|
not modelled |
11.0 |
19 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Sigma3 and sigma4 domains of RNA polymerase sigma factors
Family:Sigma4 domain |
|
|
| 72 | c4u7jB_ |
|
not modelled |
10.9 |
18 |
PDB header:ligase
Chain: B: PDB Molecule:argininosuccinate synthase;
PDBTitle: crystal structure of argininosuccinate synthase from mycobacterium2 thermoresistibile
|
|
|
| 73 | d1a04a1 |
|
not modelled |
10.9 |
14 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:C-terminal effector domain of the bipartite response regulators
Family:GerE-like (LuxR/UhpA family of transcriptional regulators) |
|
|
| 74 | d1x6oa1 |
|
not modelled |
10.6 |
18 |
Fold:SH3-like barrel
Superfamily:Translation proteins SH3-like domain
Family:eIF5a N-terminal domain-like |
|
|
| 75 | c6n1fD_ |
|
not modelled |
10.5 |
50 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:oxidoreductase, 2og-fe(ii) oxygenase family;
PDBTitle: crystal structure of oxidoreductase, 2og-fe(ii) oxygenase family, from2 burkholderia pseudomallei
|
|
|
| 76 | c2zkrq_ |
|
not modelled |
10.4 |
57 |
PDB header:ribosomal protein/rna
Chain: Q: PDB Molecule:rna expansion segment es31 part ii;
PDBTitle: structure of a mammalian ribosomal 60s subunit within an 80s complex2 obtained by docking homology models of the rna and proteins into an3 8.7 a cryo-em map
|
|
|
| 77 | c5gjqS_ |
|
not modelled |
10.3 |
17 |
PDB header:hydrolase
Chain: S: PDB Molecule:26s proteasome non-atpase regulatory subunit 3;
PDBTitle: structure of the human 26s proteasome bound to usp14-ubal
|
|
|
| 78 | c4a4fA_ |
|
not modelled |
10.2 |
20 |
PDB header:rna binding protein
Chain: A: PDB Molecule:survival of motor neuron-related-splicing factor 30;
PDBTitle: solution structure of spf30 tudor domain in complex with symmetrically2 dimethylated arginine
|
|
|
| 79 | c5a5tM_ |
|
not modelled |
9.9 |
4 |
PDB header:hydrolase
Chain: M: PDB Molecule:eukaryotic translation initiation factor 3 subunit m;
PDBTitle: structure of mammalian eif3 in the context of the 43s preinitiation2 complex
|
|
|
| 80 | c2krfB_ |
|
not modelled |
9.8 |
17 |
PDB header:transcription
Chain: B: PDB Molecule:transcriptional regulatory protein coma;
PDBTitle: nmr solution structure of the dna binding domain of competence protein2 a
|
|
|
| 81 | d1ed7a_ |
|
not modelled |
9.7 |
27 |
Fold:WW domain-like
Superfamily:Carbohydrate binding domain
Family:Carbohydrate binding domain |
|
|
| 82 | c1s1iQ_ |
|
not modelled |
9.7 |
71 |
PDB header:ribosome
Chain: Q: PDB Molecule:60s ribosomal protein l21-a;
PDBTitle: structure of the ribosomal 80s-eef2-sordarin complex from yeast2 obtained by docking atomic models for rna and protein components into3 a 11.7 a cryo-em map. this file, 1s1i, contains 60s subunit. the 40s4 ribosomal subunit is in file 1s1h.
|
|
|
| 83 | c3q1jA_ |
|
not modelled |
9.7 |
13 |
PDB header:transcription
Chain: A: PDB Molecule:phd finger protein 20;
PDBTitle: crystal structure of tudor domain 1 of human phd finger protein 20
|
|
|
| 84 | c2lxrA_ |
|
not modelled |
9.4 |
50 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nadh dehydrogenase i subunit e;
PDBTitle: solution structure of hp1264 from helicobacter pylori
|
|
|
| 85 | d1khca_ |
|
not modelled |
9.3 |
16 |
Fold:SH3-like barrel
Superfamily:Tudor/PWWP/MBT
Family:PWWP domain |
|
|
| 86 | d1l3la1 |
|
not modelled |
9.2 |
21 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:C-terminal effector domain of the bipartite response regulators
Family:GerE-like (LuxR/UhpA family of transcriptional regulators) |
|
|
| 87 | c3lpzA_ |
|
not modelled |
9.1 |
21 |
PDB header:protein transport
Chain: A: PDB Molecule:get4 (yor164c homolog);
PDBTitle: crystal structure of c. therm. get4
|
|
|
| 88 | c3llrA_ |
|
not modelled |
9.0 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:dna (cytosine-5)-methyltransferase 3a;
PDBTitle: crystal structure of the pwwp domain of human dna (cytosine-5-)-2 methyltransferase 3 alpha
|
|
|
| 89 | c3mb5A_ |
|
not modelled |
8.9 |
44 |
PDB header:transferase
Chain: A: PDB Molecule:sam-dependent methyltransferase;
PDBTitle: crystal structure of p. abyssi trna m1a58 methyltransferase in complex2 with s-adenosyl-l-methionine
|
|
|
| 90 | c4v1ak_ |
|
not modelled |
8.6 |
38 |
PDB header:ribosome
Chain: K: PDB Molecule:
PDBTitle: structure of the large subunit of the mammalian mitoribosome, part 22 of 2
|
|
|
| 91 | d1eyba_ |
|
not modelled |
8.5 |
36 |
Fold:Double-stranded beta-helix
Superfamily:RmlC-like cupins
Family:Homogentisate dioxygenase |
|
|
| 92 | c1ey2A_ |
|
not modelled |
8.5 |
36 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:homogentisate 1,2-dioxygenase;
PDBTitle: human homogentisate dioxygenase with fe(ii)
|
|
|
| 93 | c2dxcG_ |
|
not modelled |
8.5 |
63 |
PDB header:hydrolase
Chain: G: PDB Molecule:thiocyanate hydrolase subunit alpha;
PDBTitle: recombinant thiocyanate hydrolase, fully-matured form
|
|
|
| 94 | d2nlua1 |
|
not modelled |
8.3 |
18 |
Fold:SH3-like barrel
Superfamily:Tudor/PWWP/MBT
Family:PWWP domain |
|
|
| 95 | c6jqsA_ |
|
not modelled |
8.1 |
7 |
PDB header:dna binding protein
Chain: A: PDB Molecule:dna-binding response regulator;
PDBTitle: structure of transcription factor, gere
|
|
|
| 96 | d2cp5a1 |
|
not modelled |
8.1 |
15 |
Fold:SH3-like barrel
Superfamily:Cap-Gly domain
Family:Cap-Gly domain |
|
|
| 97 | c3cpfB_ |
|
not modelled |
7.9 |
9 |
PDB header:cell cycle
Chain: B: PDB Molecule:eukaryotic translation initiation factor 5a-1;
PDBTitle: crystal structure of human eukaryotic translation initiation factor2 eif5a
|
|
|
| 98 | c2d49A_ |
|
not modelled |
7.9 |
33 |
PDB header:hydrolase
Chain: A: PDB Molecule:chitinase c;
PDBTitle: solution structure of the chitin-binding domain of2 streptomyces griseus chitinase c
|
|
|
| 99 | c6nr84_ |
|
not modelled |
7.9 |
18 |
PDB header:chaperone
Chain: 4: PDB Molecule:prefoldin subunit 4;
PDBTitle: htric-hpfd class6
|
|
|