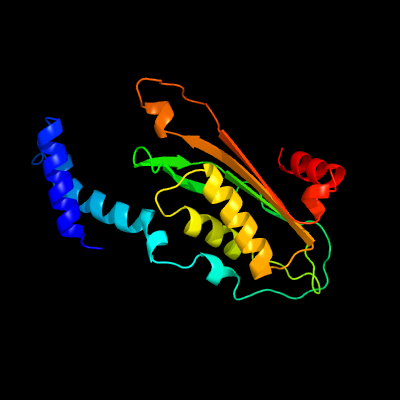
| 1 | c4uqfB_
|
|
 |
100.0 |
57 |
PDB header:hydrolase
Chain: B: PDB Molecule:gtp cyclohydrolase 1;
PDBTitle: crystal structure of listeria monocytogenes gtp cyclohydrolase i
|
|
|
|
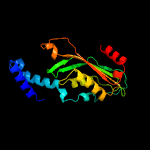
| 2 | d1wpla_
|
|
 |
100.0 |
48 |
Fold:T-fold
Superfamily:Tetrahydrobiopterin biosynthesis enzymes-like
Family:GTP cyclohydrolase I |
|
|
|
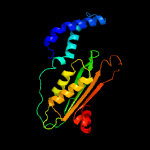
| 3 | d1wura1
|
|
 |
100.0 |
48 |
Fold:T-fold
Superfamily:Tetrahydrobiopterin biosynthesis enzymes-like
Family:GTP cyclohydrolase I |
|
|
|
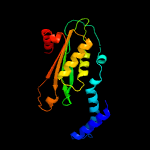
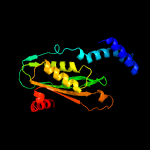
| 4 | c1wm9D_
|
|
 |
100.0 |
48 |
PDB header:hydrolase
Chain: D: PDB Molecule:gtp cyclohydrolase i;
PDBTitle: structure of gtp cyclohydrolase i from thermus thermophilus hb8
|
|
|
|
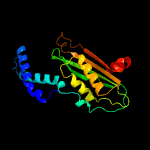
| 5 | c1is7F_
|
|
 |
100.0 |
48 |
PDB header:hydrolase/protein binding
Chain: F: PDB Molecule:gtp cyclohydrolase i;
PDBTitle: crystal structure of rat gtpchi/gfrp stimulatory complex
|
|
|
|
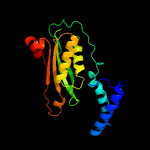
| 6 | d1a8ra_
|
|
 |
100.0 |
37 |
Fold:T-fold
Superfamily:Tetrahydrobiopterin biosynthesis enzymes-like
Family:GTP cyclohydrolase I |
|
|
|
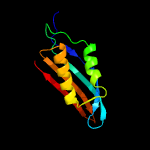
| 7 | c5jyxB_
|
|
 |
100.0 |
22 |
PDB header:transferase
Chain: B: PDB Molecule:archeaosine synthase quef-like;
PDBTitle: crystal structure of the covalent thioimide intermediate of the2 archaeosine synthase quef-like
|
|
|
|
| 8 | c4f8bE_
|
|
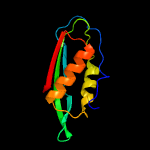 |
99.8 |
19 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:nadph-dependent 7-cyano-7-deazaguanine reductase;
PDBTitle: crystal structure of the covalent thioimide intermediate of unimodular2 nitrile reductase quef
|
|
|
|
| 9 | c3bp1A_
|
|
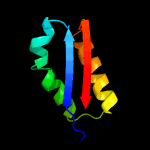 |
91.4 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nadph-dependent 7-cyano-7-deazaguanine reductase;
PDBTitle: crystal structure of putative 7-cyano-7-deazaguanine reductase quef2 from vibrio cholerae o1 biovar eltor
|
|
|
|
| 10 | c3pg6D_
|
|
 |
54.6 |
19 |
PDB header:ligase
Chain: D: PDB Molecule:e3 ubiquitin-protein ligase dtx3l;
PDBTitle: the carboxyl terminal domain of human deltex 3-like
|
|
|
|
| 11 | c6em34_
|
|
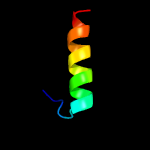 |
28.7 |
24 |
PDB header:ribosome
Chain: 4: PDB Molecule:ribosomal rna-processing protein 1;
PDBTitle: state a architectural model (nsa1-tap flag-ytm1) - visualizing the2 assembly pathway of nucleolar pre-60s ribosomes
|
|
|
|
| 12 | d1oz9a_
|
|
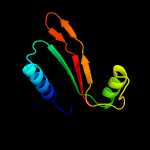 |
27.6 |
15 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Predicted metal-dependent hydrolase |
|
|
|
| 13 | d1o6ca_
|
|
 |
27.0 |
12 |
Fold:UDP-Glycosyltransferase/glycogen phosphorylase
Superfamily:UDP-Glycosyltransferase/glycogen phosphorylase
Family:UDP-N-acetylglucosamine 2-epimerase |
|
|
|
| 14 | c3iraA_
|
|
 |
26.8 |
14 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:conserved protein;
PDBTitle: the crystal structure of one domain of the conserved protein from2 methanosarcina mazei go1
|
|
|
|
| 15 | d1dpta_
|
|
 |
25.5 |
20 |
Fold:Tautomerase/MIF
Superfamily:Tautomerase/MIF
Family:MIF-related |
|
|
|
| 16 | c2xczA_
|
|
 |
24.5 |
13 |
PDB header:immune system
Chain: A: PDB Molecule:possible atls1-like light-inducible protein;
PDBTitle: crystal structure of macrophage migration inhibitory factor homologue2 from prochlorococcus marinus
|
|
|
|
| 17 | c3ugsB_
|
|
 |
23.9 |
19 |
PDB header:transferase
Chain: B: PDB Molecule:undecaprenyl pyrophosphate synthase;
PDBTitle: crystal structure of a probable undecaprenyl diphosphate synthase2 (upps) from campylobacter jejuni
|
|
|
|
| 18 | d1uiza_
|
|
 |
23.8 |
17 |
Fold:Tautomerase/MIF
Superfamily:Tautomerase/MIF
Family:MIF-related |
|
|
|
| 19 | c4g6cA_
|
|
 |
22.6 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-hexosaminidase 1;
PDBTitle: crystal structure of beta-hexosaminidase 1 from burkholderia2 cenocepacia j2315
|
|
|
|
| 20 | c2d2rA_
|
|
 |
21.9 |
32 |
PDB header:transferase
Chain: A: PDB Molecule:undecaprenyl pyrophosphate synthase;
PDBTitle: crystal structure of helicobacter pylori undecaprenyl pyrophosphate2 synthase
|
|
|
|
| 21 | c5l0uA_ |
|
not modelled |
21.7 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:protein o-glucosyltransferase 1;
PDBTitle: human poglut1 in complex with egf(+) and udp-phosphono-glucose
|
|
|
| 22 | c5z3gX_ |
|
not modelled |
19.4 |
25 |
PDB header:ribosome
Chain: X: PDB Molecule:ribosomal rna-processing protein 1;
PDBTitle: cryo-em structure of a nucleolar pre-60s ribosome (rpf1-tap)
|
|
|
| 23 | c5enzA_ |
|
not modelled |
18.9 |
16 |
PDB header:isomerase
Chain: A: PDB Molecule:udp-glcnac 2-epimerase;
PDBTitle: s. aureus mnaa-udp co-structure
|
|
|
| 24 | d2veaa1 |
|
not modelled |
18.5 |
7 |
Fold:Profilin-like
Superfamily:GAF domain-like
Family:GAF domain |
|
|
| 25 | c1jp3A_ |
|
not modelled |
18.0 |
48 |
PDB header:transferase
Chain: A: PDB Molecule:undecaprenyl pyrophosphate synthase;
PDBTitle: structure of e.coli undecaprenyl pyrophosphate synthase
|
|
|
| 26 | c2l9vA_ |
|
not modelled |
17.9 |
19 |
PDB header:rna binding protein
Chain: A: PDB Molecule:pre-mrna-processing factor 40 homolog a;
PDBTitle: nmr structure of the ff domain l24a mutant's folding transition state
|
|
|
| 27 | c6emfA_ |
|
not modelled |
17.0 |
25 |
PDB header:ribosome
Chain: A: PDB Molecule:g0s4m2;
PDBTitle: crystal structure of rrp1 from chaetomium thermophilum in space group2 c2
|
|
|
| 28 | d1vqoj1 |
|
not modelled |
16.8 |
26 |
Fold:Ribosomal protein L13
Superfamily:Ribosomal protein L13
Family:Ribosomal protein L13 |
|
|
| 29 | c3zf7Y_ |
|
not modelled |
16.6 |
13 |
PDB header:ribosome
Chain: Y: PDB Molecule:60s ribosomal protein l24, putative;
PDBTitle: high-resolution cryo-electron microscopy structure of the trypanosoma2 brucei ribosome
|
|
|
| 30 | c5xk9F_ |
|
not modelled |
15.2 |
24 |
PDB header:transferase
Chain: F: PDB Molecule:undecaprenyl diphosphate synthase;
PDBTitle: crystal structure of isosesquilavandulyl diphosphate synthase from2 streptomyces sp. strain cnh-189 in complex with gspp and dmapp
|
|
|
| 31 | c5gukA_ |
|
not modelled |
15.1 |
28 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:cyclolavandulyl diphosphate synthase;
PDBTitle: crystal structure of apo form of cyclolavandulyl diphosphate synthase2 (clds) from streptomyces sp. cl190
|
|
|
| 32 | d1f75a_ |
|
not modelled |
15.0 |
20 |
Fold:Undecaprenyl diphosphate synthase
Superfamily:Undecaprenyl diphosphate synthase
Family:Undecaprenyl diphosphate synthase |
|
|
| 33 | d2nn6f2 |
|
not modelled |
15.0 |
29 |
Fold:Ribonuclease PH domain 2-like
Superfamily:Ribonuclease PH domain 2-like
Family:Ribonuclease PH domain 2-like |
|
|
| 34 | c6nuwF_ |
|
not modelled |
14.8 |
22 |
PDB header:cell cycle
Chain: F: PDB Molecule:inner kinetochore subunit okp1;
PDBTitle: yeast ctf19 complex
|
|
|
| 35 | d1j09a2 |
|
not modelled |
13.8 |
35 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Nucleotidylyl transferase
Family:Class I aminoacyl-tRNA synthetases (RS), catalytic domain |
|
|
| 36 | d1ueha_ |
|
not modelled |
13.8 |
48 |
Fold:Undecaprenyl diphosphate synthase
Superfamily:Undecaprenyl diphosphate synthase
Family:Undecaprenyl diphosphate synthase |
|
|
| 37 | c5hxpA_ |
|
not modelled |
12.7 |
36 |
PDB header:transferase
Chain: A: PDB Molecule:(2z,6z)-farnesyl diphosphate synthase, chloroplastic;
PDBTitle: crystal structure of z,z-farnesyl diphosphate synthase (d71m, e75a and2 h103y mutants) complexed with ipp
|
|
|
| 38 | c5f86A_ |
|
not modelled |
12.3 |
12 |
PDB header:transferase/hydrolase
Chain: A: PDB Molecule:o-glucosyltransferase rumi;
PDBTitle: crystal structure of drosophila poglut1 (rumi) complexed with its2 substrate protein (egf repeat)
|
|
|
| 39 | c2o90A_ |
|
not modelled |
12.3 |
18 |
PDB header:lyase
Chain: A: PDB Molecule:dihydroneopterin aldolase;
PDBTitle: atomic resolution crystal structure of e.coli dihydroneopterin2 aldolase in complex with neopterin
|
|
|
| 40 | c5hg1A_ |
|
not modelled |
12.0 |
16 |
PDB header:transferase/transferase inhibitor
Chain: A: PDB Molecule:hexokinase-2;
PDBTitle: crystal structure of human hexokinase 2 with cmpd 1, a c-2-substituted2 glucosamine
|
|
|
| 41 | c3f6nA_ |
|
not modelled |
11.9 |
21 |
PDB header:viral protein, dna-binding protein
Chain: A: PDB Molecule:virion-associated protein;
PDBTitle: crystal structure of the virion-associated protein p3 from2 caulimovirus
|
|
|
| 42 | c3k4tB_ |
|
not modelled |
11.6 |
21 |
PDB header:viral protein, dna-binding protein
Chain: B: PDB Molecule:virion-associated protein;
PDBTitle: crystal structure of the virion-associated protein p3 from2 caulimovirus
|
|
|
| 43 | c5ljiA_ |
|
not modelled |
11.6 |
38 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:flavodoxin;
PDBTitle: streptococcus pneumonia tigr4 flavodoxin: structural and biophysical2 characterization of a novel drug target
|
|
|
| 44 | c2lmkA_ |
|
not modelled |
11.5 |
37 |
PDB header:signaling protein
Chain: A: PDB Molecule:exocrine gland-secreting peptide 1;
PDBTitle: solution structure of mouse pheromone esp1
|
|
|
| 45 | c5bzaA_ |
|
not modelled |
11.4 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-n-acetylhexosaminidase;
PDBTitle: crystal structure of cbsa from thermotoga neapolitana
|
|
|
| 46 | c3aiiA_ |
|
not modelled |
11.4 |
31 |
PDB header:ligase
Chain: A: PDB Molecule:glutamyl-trna synthetase;
PDBTitle: archaeal non-discriminating glutamyl-trna synthetase from2 methanothermobacter thermautotrophicus
|
|
|
| 47 | d1vhxa_ |
|
not modelled |
11.3 |
27 |
Fold:Ribonuclease H-like motif
Superfamily:Ribonuclease H-like
Family:Putative Holliday junction resolvase RuvX |
|
|
| 48 | c3tevA_ |
|
not modelled |
11.2 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:glycosyl hyrolase, family 3;
PDBTitle: the crystal structure of glycosyl hydrolase from deinococcus2 radiodurans r1
|
|
|
| 49 | c5odcD_ |
|
not modelled |
11.1 |
18 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:methyl-viologen reducing hydrogenase subunit d;
PDBTitle: heterodisulfide reductase / [nife]-hydrogenase complex from2 methanothermococcus thermolithotrophicus at 2.3 a resolution
|
|
|
| 50 | d3eeqa1 |
|
not modelled |
11.0 |
8 |
Fold:CobE/GbiG C-terminal domain-like
Superfamily:CobE/GbiG C-terminal domain-like
Family:CobE/GbiG C-terminal domain-like |
|
|
| 51 | c5hc7A_ |
|
not modelled |
10.9 |
28 |
PDB header:transferase/transferase inhibitor
Chain: A: PDB Molecule:prenyltransference for protein;
PDBTitle: crystal structure of lavandulyl diphosphate synthase from lavandula x2 intermedia in complex with s-thiolo-isopentenyldiphosphate
|
|
|
| 52 | c3femB_ |
|
not modelled |
10.8 |
32 |
PDB header:biosynthetic protein, transferase
Chain: B: PDB Molecule:pyridoxine biosynthesis protein snz1;
PDBTitle: structure of the synthase subunit pdx1.1 (snz1) of plp synthase from2 saccharomyces cerevisiae
|
|
|
| 53 | d1b9la_ |
|
not modelled |
10.7 |
22 |
Fold:T-fold
Superfamily:Tetrahydrobiopterin biosynthesis enzymes-like
Family:DHN aldolase/epimerase |
|
|
| 54 | c4m1aB_ |
|
not modelled |
10.6 |
10 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:hypothetical protein;
PDBTitle: crystal structure of a domain of unknown function (duf1904) from2 sebaldella termitidis atcc 33386
|
|
|
| 55 | c6c0dA_ |
|
not modelled |
10.6 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:amidase, hydantoinase/carbamoylase family;
PDBTitle: crystal structure of an amidase (hydantoinase/carbamoylase family)2 from burkholderia phymatum
|
|
|
| 56 | c3fwtA_ |
|
not modelled |
10.5 |
10 |
PDB header:cytokine
Chain: A: PDB Molecule:macrophage migration inhibitory factor-like
PDBTitle: crystal structure of leishmania major mif2
|
|
|
| 57 | c4v19N_ |
|
not modelled |
10.0 |
20 |
PDB header:ribosome
Chain: N: PDB Molecule:mitoribosomal protein ul13m, mrpl13;
PDBTitle: structure of the large subunit of the mammalian mitoribosome, part 12 of 2
|
|
|
| 58 | c6fsiA_ |
|
not modelled |
9.9 |
14 |
PDB header:electron transport
Chain: A: PDB Molecule:flavodoxin;
PDBTitle: crystal structure of semiquinone flavodoxin 1 from bacillus cereus2 (1.32 a resolution)
|
|
|
| 59 | d1u8va1 |
|
not modelled |
9.9 |
19 |
Fold:Bromodomain-like
Superfamily:Acyl-CoA dehydrogenase C-terminal domain-like
Family:Medium chain acyl-CoA dehydrogenase-like, C-terminal domain |
|
|
| 60 | c6gaqB_ |
|
not modelled |
9.7 |
38 |
PDB header:electron transport
Chain: B: PDB Molecule:flavodoxin;
PDBTitle: crystal structure of oxidised flavodoxin 2 from bacillus cereus
|
|
|
| 61 | d1nzja_ |
|
not modelled |
9.7 |
31 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Nucleotidylyl transferase
Family:Class I aminoacyl-tRNA synthetases (RS), catalytic domain |
|
|
| 62 | c5w6yB_ |
|
not modelled |
9.7 |
12 |
PDB header:biosynthetic protein,isomerase
Chain: B: PDB Molecule:chorismate mutase;
PDBTitle: physcomitrella patens chorismate mutase
|
|
|
| 63 | c3lk6A_ |
|
not modelled |
9.5 |
20 |
PDB header:hydrolase
Chain: A: PDB Molecule:lipoprotein ybbd;
PDBTitle: beta-n-hexosaminidase n318d mutant (ybbd_n318d) from bacillus subtilis
|
|
|
| 64 | c3sqlB_ |
|
not modelled |
9.5 |
18 |
PDB header:hydrolase
Chain: B: PDB Molecule:glycosyl hydrolase family 3;
PDBTitle: crystal structure of glycoside hydrolase from synechococcus
|
|
|
| 65 | c6hcyA_ |
|
not modelled |
9.2 |
14 |
PDB header:membrane protein
Chain: A: PDB Molecule:metalloreductase steap4;
PDBTitle: human steap4 bound to nadp, fad, heme and fe(iii)-nta.
|
|
|
| 66 | d1j3aa_ |
|
not modelled |
9.2 |
26 |
Fold:Ribosomal protein L13
Superfamily:Ribosomal protein L13
Family:Ribosomal protein L13 |
|
|
| 67 | c2zbtB_ |
|
not modelled |
9.2 |
32 |
PDB header:lyase
Chain: B: PDB Molecule:pyridoxal biosynthesis lyase pdxs;
PDBTitle: crystal structure of pyridoxine biosynthesis protein from thermus2 thermophilus hb8
|
|
|
| 68 | d1u9da_ |
|
not modelled |
9.1 |
22 |
Fold:Tautomerase/MIF
Superfamily:Tautomerase/MIF
Family:VC0714-like |
|
|
| 69 | c2kduB_ |
|
not modelled |
9.0 |
67 |
PDB header:metal binding protein/exocytosis
Chain: B: PDB Molecule:protein unc-13 homolog a;
PDBTitle: structural basis of the munc13-1/ca2+-calmodulin2 interaction: a novel 1-26 calmodulin binding motif with a3 bipartite binding mode
|
|
|
| 70 | c3gacD_ |
|
not modelled |
8.9 |
16 |
PDB header:cytokine
Chain: D: PDB Molecule:macrophage migration inhibitory factor-like
PDBTitle: structure of mif with hpp
|
|
|
| 71 | c3d5bN_ |
|
not modelled |
8.8 |
29 |
PDB header:ribosome
Chain: N: PDB Molecule:50s ribosomal protein l13;
PDBTitle: structural basis for translation termination on the 70s ribosome. this2 file contains the 50s subunit of one 70s ribosome. the entire crystal3 structure contains two 70s ribosomes as described in remark 400.
|
|
|
| 72 | d1fima_ |
|
not modelled |
8.8 |
17 |
Fold:Tautomerase/MIF
Superfamily:Tautomerase/MIF
Family:MIF-related |
|
|
| 73 | c4firB_ |
|
not modelled |
8.7 |
32 |
PDB header:lyase
Chain: B: PDB Molecule:pyridoxal biosynthesis lyase pdxs;
PDBTitle: crystal structure of pyridoxal biosynthesis lyase pdxs from pyrococcus
|
|
|
| 74 | d5csma_ |
|
not modelled |
8.7 |
24 |
Fold:Chorismate mutase II
Superfamily:Chorismate mutase II
Family:Allosteric chorismate mutase |
|
|
| 75 | d1nbua_ |
|
not modelled |
8.6 |
22 |
Fold:T-fold
Superfamily:Tetrahydrobiopterin biosynthesis enzymes-like
Family:DHN aldolase/epimerase |
|
|
| 76 | c6gcs8_ |
|
not modelled |
8.5 |
33 |
PDB header:oxidoreductase
Chain: 8: PDB Molecule:nb8m subunit;
PDBTitle: cryo-em structure of respiratory complex i from yarrowia lipolytica
|
|
|
| 77 | d1vmea1 |
|
not modelled |
8.5 |
14 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:Flavodoxin-related |
|
|
| 78 | c3b64A_ |
|
not modelled |
8.5 |
15 |
PDB header:cytokine
Chain: A: PDB Molecule:macrophage migration inhibitory factor-like
PDBTitle: macrophage migration inhibitory factor (mif) from2 /leishmania major
|
|
|
| 79 | c5t58N_ |
|
not modelled |
8.5 |
19 |
PDB header:cell cycle
Chain: N: PDB Molecule:klla0c15939p;
PDBTitle: structure of the mind complex shows a regulatory focus of yeast2 kinetochore assembly
|
|
|
| 80 | c2vfwB_ |
|
not modelled |
8.5 |
22 |
PDB header:transferase
Chain: B: PDB Molecule:short-chain z-isoprenyl diphosphate synthetase;
PDBTitle: rv1086 native
|
|
|
| 81 | d1d8ja_ |
|
not modelled |
8.4 |
21 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:The central core domain of TFIIE beta |
|
|
| 82 | c4dh4A_ |
|
not modelled |
8.3 |
16 |
PDB header:isomerase
Chain: A: PDB Molecule:mif;
PDBTitle: macrophage migration inhibitory factor toxoplasma gondii
|
|
|
| 83 | c2nv2U_ |
|
not modelled |
8.2 |
34 |
PDB header:lyase/transferase
Chain: U: PDB Molecule:pyridoxal biosynthesis lyase pdxs;
PDBTitle: structure of the plp synthase complex pdx1/2 (yaad/e) from bacillus2 subtilis
|
|
|
| 84 | d1v29a_ |
|
not modelled |
8.2 |
18 |
Fold:Nitrile hydratase alpha chain
Superfamily:Nitrile hydratase alpha chain
Family:Nitrile hydratase alpha chain |
|
|
| 85 | d1cmca_ |
|
not modelled |
8.2 |
21 |
Fold:Ribbon-helix-helix
Superfamily:Ribbon-helix-helix
Family:Met repressor, MetJ (MetR) |
|
|
| 86 | c3ngvA_ |
|
not modelled |
8.2 |
14 |
PDB header:transport protein
Chain: A: PDB Molecule:d7 protein;
PDBTitle: crystal structure of anst-d7l1
|
|
|
| 87 | d2o16a3 |
|
not modelled |
8.1 |
15 |
Fold:CBS-domain pair
Superfamily:CBS-domain pair
Family:CBS-domain pair |
|
|
| 88 | d1lfpa_ |
|
not modelled |
8.1 |
36 |
Fold:YebC-like
Superfamily:YebC-like
Family:YebC-like |
|
|
| 89 | c3wo8B_ |
|
not modelled |
8.1 |
19 |
PDB header:hydrolase
Chain: B: PDB Molecule:beta-n-acetylglucosaminidase;
PDBTitle: crystal structure of the beta-n-acetylglucosaminidase from thermotoga2 maritima
|
|
|
| 90 | c3cf5G_ |
|
not modelled |
7.9 |
23 |
PDB header:ribosome/antibiotic
Chain: G: PDB Molecule:50s ribosomal protein l13;
PDBTitle: thiopeptide antibiotic thiostrepton bound to the large ribosomal2 subunit of deinococcus radiodurans
|
|
|
| 91 | d2zjrg1 |
|
not modelled |
7.9 |
23 |
Fold:Ribosomal protein L13
Superfamily:Ribosomal protein L13
Family:Ribosomal protein L13 |
|
|
| 92 | c4h3sA_ |
|
not modelled |
7.7 |
11 |
PDB header:ligase
Chain: A: PDB Molecule:glutamine-trna ligase;
PDBTitle: the structure of glutaminyl-trna synthetase from saccharomyces2 cerevisiae
|
|
|
| 93 | c1zn1L_ |
|
not modelled |
7.7 |
33 |
PDB header:biosynthetic/structural protein/rna
Chain: L: PDB Molecule:30s ribosomal protein s12;
PDBTitle: coordinates of rrf fitted into cryo-em map of the 70s post-termination2 complex
|
|
|
| 94 | c2k29A_ |
|
not modelled |
7.6 |
24 |
PDB header:transcription
Chain: A: PDB Molecule:antitoxin relb;
PDBTitle: structure of the dbd domain of e. coli antitoxin relb
|
|
|
| 95 | d2d28c1 |
|
not modelled |
7.6 |
15 |
Fold:Alpha-lytic protease prodomain-like
Superfamily:EspE N-terminal domain-like
Family:GSPII protein E N-terminal domain-like |
|
|
| 96 | c2yzrB_ |
|
not modelled |
7.6 |
37 |
PDB header:lyase
Chain: B: PDB Molecule:pyridoxal biosynthesis lyase pdxs;
PDBTitle: crystal structure of pyridoxine biosynthesis protein from2 methanocaldococcus jannaschii
|
|
|
| 97 | c4rn7A_ |
|
not modelled |
7.5 |
30 |
PDB header:hydrolase
Chain: A: PDB Molecule:n-acetylmuramoyl-l-alanine amidase;
PDBTitle: the crystal structure of n-acetylmuramoyl-l-alanine amidase from2 clostridium difficile 630
|
|
|
| 98 | c3uv1B_ |
|
not modelled |
7.5 |
22 |
PDB header:allergen
Chain: B: PDB Molecule:der f 7 allergen;
PDBTitle: crystal structure a major allergen from dust mite
|
|
|
| 99 | c3hlyA_ |
|
not modelled |
7.4 |
24 |
PDB header:flavoprotein
Chain: A: PDB Molecule:flavodoxin-like domain;
PDBTitle: crystal structure of the flavodoxin-like domain from synechococcus sp2 q5mzp6_synp6 protein. northeast structural genomics consortium target3 snr135d.
|
|
|