| 1 |
|
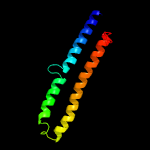
PDB 3gvm chain A
Region: 1 - 98
Aligned: 94
Modelled: 98
Confidence: 97.7%
Identity: 13%
PDB header:viral protein
Chain: A: PDB Molecule:putative uncharacterized protein sag1039;
PDBTitle: structure of the homodimeric wxg-100 family protein from streptococcus2 agalactiae
Phyre2
| 2 |
|
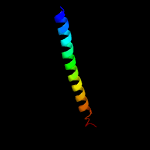
PDB 4iog chain D
Region: 1 - 90
Aligned: 86
Modelled: 90
Confidence: 97.7%
Identity: 16%
PDB header:unknown function
Chain: D: PDB Molecule:secreted protein esxb;
PDBTitle: the crystal structure of a secreted protein esxb (wild-type, in p212 space group) from bacillus anthracis str. sterne
Phyre2
| 3 |
|
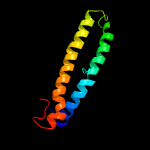
PDB 1wa8 chain A domain 1
Region: 4 - 98
Aligned: 92
Modelled: 95
Confidence: 97.5%
Identity: 13%
Fold: Ferritin-like
Superfamily: EsxAB dimer-like
Family: ESAT-6 like
Phyre2
| 4 |
|
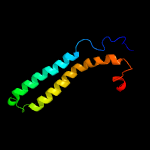
PDB 3zbh chain C
Region: 8 - 100
Aligned: 90
Modelled: 93
Confidence: 97.3%
Identity: 12%
PDB header:unknown function
Chain: C: PDB Molecule:esxa;
PDBTitle: geobacillus thermodenitrificans esxa crystal form i
Phyre2
| 5 |
|
PDB 2vs0 chain B
Region: 5 - 91
Aligned: 84
Modelled: 87
Confidence: 97.1%
Identity: 10%
PDB header:cell invasion
Chain: B: PDB Molecule:virulence factor esxa;
PDBTitle: structural analysis of homodimeric staphylococcal aureus2 virulence factor esxa
Phyre2
| 6 |
|
PDB 4lws chain A
Region: 4 - 101
Aligned: 95
Modelled: 98
Confidence: 96.5%
Identity: 13%
PDB header:unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: esxa : esxb (semet) hetero-dimer from thermomonospora curvata
Phyre2
| 7 |
|
PDB 4lws chain B
Region: 1 - 88
Aligned: 85
Modelled: 88
Confidence: 96.0%
Identity: 11%
PDB header:unknown function
Chain: B: PDB Molecule:uncharacterized protein;
PDBTitle: esxa : esxb (semet) hetero-dimer from thermomonospora curvata
Phyre2
| 8 |
|
PDB 4wj2 chain A
Region: 60 - 98
Aligned: 39
Modelled: 39
Confidence: 91.7%
Identity: 21%
PDB header:unknown function
Chain: A: PDB Molecule:antigen mtb48;
PDBTitle: mycobacterial protein
Phyre2
| 9 |
|
PDB 1wa8 chain B domain 1
Region: 4 - 96
Aligned: 90
Modelled: 93
Confidence: 90.9%
Identity: 10%
Fold: Ferritin-like
Superfamily: EsxAB dimer-like
Family: ESAT-6 like
Phyre2
| 10 |
|
PDB 2kg7 chain B
Region: 3 - 100
Aligned: 95
Modelled: 95
Confidence: 83.8%
Identity: 12%
PDB header:unknown function
Chain: B: PDB Molecule:esat-6-like protein esxh;
PDBTitle: structure and features of the complex formed by the tuberculosis2 virulence factors rv0287 and rv0288
Phyre2
| 11 |
|
PDB 4i0x chain A
Region: 10 - 87
Aligned: 75
Modelled: 78
Confidence: 63.0%
Identity: 16%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:esat-6-like protein mab_3112;
PDBTitle: crystal structure of the mycobacterum abscessus esxef (mab_3112-2 mab_3113) complex
Phyre2
| 12 |
|
PDB 4xy3 chain A
Region: 60 - 92
Aligned: 33
Modelled: 33
Confidence: 60.5%
Identity: 30%
PDB header:protein transport
Chain: A: PDB Molecule:esx-1 secretion-associated protein espb;
PDBTitle: structure of esx-1 secreted protein espb
Phyre2
| 13 |
|
PDB 2kg7 chain A
Region: 53 - 91
Aligned: 39
Modelled: 39
Confidence: 12.0%
Identity: 21%
PDB header:unknown function
Chain: A: PDB Molecule:uncharacterized protein esxg (pe family protein);
PDBTitle: structure and features of the complex formed by the tuberculosis2 virulence factors rv0287 and rv0288
Phyre2
| 14 |
|
PDB 3jyw chain M
Region: 47 - 86
Aligned: 39
Modelled: 40
Confidence: 10.9%
Identity: 15%
PDB header:ribosome
Chain: M: PDB Molecule:60s ribosomal protein l16(a);
PDBTitle: structure of the 60s proteins for eukaryotic ribosome based on cryo-em2 map of thermomyces lanuginosus ribosome at 8.9a resolution
Phyre2
| 15 |
|
PDB 2ixs chain B
Region: 38 - 58
Aligned: 20
Modelled: 21
Confidence: 7.8%
Identity: 30%
PDB header:hydrolase
Chain: B: PDB Molecule:sdai restriction endonuclease;
PDBTitle: structure of sdai restriction endonuclease
Phyre2
| 16 |
|
PDB 1zcz chain A domain 1
Region: 80 - 90
Aligned: 11
Modelled: 11
Confidence: 6.6%
Identity: 45%
Fold: Methylglyoxal synthase-like
Superfamily: Methylglyoxal synthase-like
Family: Inosicase
Phyre2
| 17 |
|
PDB 6nko chain A
Region: 80 - 90
Aligned: 11
Modelled: 11
Confidence: 6.4%
Identity: 45%
PDB header:unknown function
Chain: A: PDB Molecule:forh;
PDBTitle: crystal structure of forh
Phyre2
| 18 |
|
PDB 5jx6 chain C
Region: 5 - 47
Aligned: 43
Modelled: 43
Confidence: 6.4%
Identity: 7%
PDB header:hydrolase
Chain: C: PDB Molecule:glucanase;
PDBTitle: gh6 orpinomyces sp. y102 enzyme
Phyre2
| 19 |
|
PDB 1pkx chain A domain 1
Region: 80 - 90
Aligned: 11
Modelled: 11
Confidence: 6.1%
Identity: 45%
Fold: Methylglyoxal synthase-like
Superfamily: Methylglyoxal synthase-like
Family: Inosicase
Phyre2