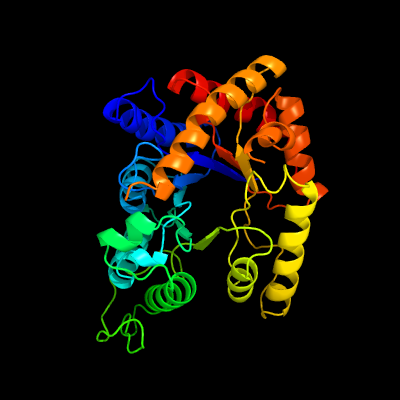
| 1 | d1luca_
|
|
 |
100.0 |
22 |
Fold:TIM beta/alpha-barrel
Superfamily:Bacterial luciferase-like
Family:Bacterial luciferase (alkanal monooxygenase) |
|
|
|
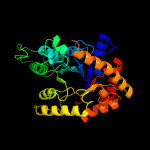
| 2 | c2wgkA_
|
|
 |
100.0 |
25 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:3,6-diketocamphane 1,6 monooxygenase;
PDBTitle: type ii baeyer-villiger monooxygenase oxygenating2 constituent of 3,6-diketocamphane 1,6 monooxygenase from3 pseudomonas putida
|
|
|
|
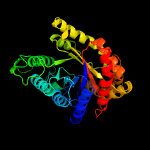
| 3 | c2i7gA_
|
|
 |
100.0 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:monooxygenase;
PDBTitle: crystal structure of monooxygenase from agrobacterium tumefaciens
|
|
|
|
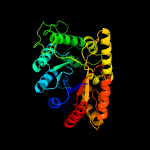
| 4 | d1lucb_
|
|
 |
100.0 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Bacterial luciferase-like
Family:Bacterial luciferase (alkanal monooxygenase) |
|
|
|
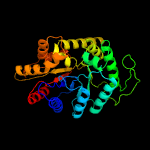
| 5 | c6friD_
|
|
 |
100.0 |
17 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:alkanal monooxygenase beta chain;
PDBTitle: structure of luxb from photobacterium leiognathi
|
|
|
|
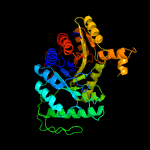
| 6 | c5tlcA_
|
|
 |
100.0 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dibenzothiophene desulfurization enzyme a;
PDBTitle: crystal structure of bdsa from bacillus subtilis wu-s2b
|
|
|
|
| 7 | d1tvla_
|
|
 |
100.0 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Bacterial luciferase-like
Family:Ssud-like monoxygenases |
|
|
|
| 8 | c1tvlA_
|
|
 |
100.0 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:protein ytnj;
PDBTitle: structure of ytnj from bacillus subtilis
|
|
|
|
| 9 | c3raoB_
|
|
 |
100.0 |
16 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:putative luciferase-like monooxygenase;
PDBTitle: crystal structure of the luciferase-like monooxygenase from bacillus2 cereus atcc 10987.
|
|
|
|
| 10 | c3sdoB_
|
|
 |
100.0 |
20 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:nitrilotriacetate monooxygenase;
PDBTitle: structure of a nitrilotriacetate monooxygenase from burkholderia2 pseudomallei
|
|
|
|
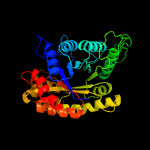
| 11 | d1ezwa_
|
|
 |
100.0 |
22 |
Fold:TIM beta/alpha-barrel
Superfamily:Bacterial luciferase-like
Family:F420 dependent oxidoreductases |
|
|
|
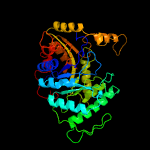
| 12 | c3b9nB_
|
|
 |
100.0 |
21 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:alkane monoxygenase;
PDBTitle: crystal structure of long-chain alkane monooxygenase (lada)
|
|
|
|
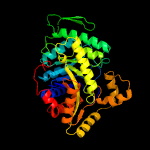
| 13 | c5w4zA_
|
|
 |
100.0 |
20 |
PDB header:flavoprotein
Chain: A: PDB Molecule:riboflavin lyase;
PDBTitle: crystal structure of riboflavin lyase (rcae) with modified fmn and2 substrate riboflavin
|
|
|
|
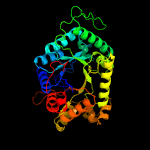
| 14 | c5dqpA_
|
|
 |
100.0 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:edta monooxygenase;
PDBTitle: edta monooxygenase (emoa) from chelativorans sp. bnc1
|
|
|
|
| 15 | c1z69D_
|
|
 |
100.0 |
19 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:coenzyme f420-dependent n(5),n(10)-
PDBTitle: crystal structure of methylenetetrahydromethanopterin2 reductase (mer) in complex with coenzyme f420
|
|
|
|
| 16 | d1nqka_
|
|
 |
100.0 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Bacterial luciferase-like
Family:Ssud-like monoxygenases |
|
|
|
| 17 | c5wanA_
|
|
 |
100.0 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:pyrimidine monooxygenase ruta;
PDBTitle: crystal structure of a flavoenzyme ruta in the pyrimidine catabolic2 pathway
|
|
|
|
| 18 | c6ak1B_
|
|
 |
100.0 |
22 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:dimethyl-sulfide monooxygenase;
PDBTitle: crystal structure of dmoa from hyphomicrobium sulfonivorans
|
|
|
|
| 19 | d1f07a_
|
|
 |
100.0 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:Bacterial luciferase-like
Family:F420 dependent oxidoreductases |
|
|
|
| 20 | c3c8nB_
|
|
 |
100.0 |
22 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:probable f420-dependent glucose-6-phosphate dehydrogenase
PDBTitle: crystal structure of apo-fgd1 from mycobacterium tuberculosis
|
|
|
|
| 21 | d1rhca_ |
|
not modelled |
100.0 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Bacterial luciferase-like
Family:F420 dependent oxidoreductases |
|
|
| 22 | c2b81D_ |
|
not modelled |
100.0 |
18 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:luciferase-like monooxygenase;
PDBTitle: crystal structure of the luciferase-like monooxygenase from bacillus2 cereus
|
|
|
| 23 | d1nfpa_ |
|
not modelled |
100.0 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Bacterial luciferase-like
Family:Non-fluorescent flavoprotein (luxF, FP390) |
|
|
| 24 | d1fvpa_ |
|
not modelled |
99.7 |
11 |
Fold:TIM beta/alpha-barrel
Superfamily:Bacterial luciferase-like
Family:Non-fluorescent flavoprotein (luxF, FP390) |
|
|
| 25 | c4xkyC_ |
|
not modelled |
88.2 |
10 |
PDB header:lyase
Chain: C: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: structure of dihydrodipicolinate synthase from the commensal bacterium2 bacteroides thetaiotaomicron at 2.1 a resolution
|
|
|
| 26 | c2infB_ |
|
not modelled |
83.9 |
6 |
PDB header:lyase
Chain: B: PDB Molecule:uroporphyrinogen decarboxylase;
PDBTitle: crystal structure of uroporphyrinogen decarboxylase from bacillus2 subtilis
|
|
|
| 27 | d1r3sa_ |
|
not modelled |
79.0 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:UROD/MetE-like
Family:Uroporphyrinogen decarboxylase, UROD |
|
|
| 28 | c4exqA_ |
|
not modelled |
78.8 |
11 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:uroporphyrinogen decarboxylase;
PDBTitle: crystal structure of uroporphyrinogen decarboxylase (upd) from2 burkholderia thailandensis e264
|
|
|
| 29 | c2ejaB_ |
|
not modelled |
78.7 |
11 |
PDB header:lyase
Chain: B: PDB Molecule:uroporphyrinogen decarboxylase;
PDBTitle: crystal structure of uroporphyrinogen decarboxylase from2 aquifex aeolicus
|
|
|
| 30 | c3b4uB_ |
|
not modelled |
77.0 |
14 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from agrobacterium2 tumefaciens str. c58
|
|
|
| 31 | d1j93a_ |
|
not modelled |
74.2 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:UROD/MetE-like
Family:Uroporphyrinogen decarboxylase, UROD |
|
|
| 32 | c1jpkA_ |
|
not modelled |
73.8 |
13 |
PDB header:lyase
Chain: A: PDB Molecule:uroporphyrinogen decarboxylase;
PDBTitle: gly156asp mutant of human urod, human uroporphyrinogen iii2 decarboxylase
|
|
|
| 33 | c3cyvA_ |
|
not modelled |
72.5 |
13 |
PDB header:lyase
Chain: A: PDB Molecule:uroporphyrinogen decarboxylase;
PDBTitle: crystal structure of uroporphyrinogen decarboxylase from2 shigella flexineri: new insights into its catalytic3 mechanism
|
|
|
| 34 | c3qy6A_ |
|
not modelled |
72.1 |
24 |
PDB header:hydrolase
Chain: A: PDB Molecule:tyrosine-protein phosphatase ywqe;
PDBTitle: crystal structures of ywqe from bacillus subtilis and cpsb from2 streptococcus pneumoniae, unique metal-dependent tyrosine3 phosphatases
|
|
|
| 35 | c4uxdC_ |
|
not modelled |
62.7 |
8 |
PDB header:lyase
Chain: C: PDB Molecule:2-dehydro-3-deoxy-d-gluconate/2-dehydro-3-deoxy-
PDBTitle: 2-keto 3-deoxygluconate aldolase from picrophilus torridus
|
|
|
| 36 | c3d0cB_ |
|
not modelled |
62.5 |
14 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from oceanobacillus2 iheyensis at 1.9 a resolution
|
|
|
| 37 | c2w1vA_ |
|
not modelled |
59.5 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:nitrilase homolog 2;
PDBTitle: crystal structure of mouse nitrilase-2 at 1.4a resolution
|
|
|
| 38 | c4zr8B_ |
|
not modelled |
59.4 |
11 |
PDB header:lyase
Chain: B: PDB Molecule:uroporphyrinogen decarboxylase;
PDBTitle: structure of uroporphyrinogen decarboxylase from acinetobacter2 baumannii
|
|
|
| 39 | c5ud6B_ |
|
not modelled |
58.7 |
14 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dhdps from cyanidioschyzon merolae with lysine2 bound
|
|
|
| 40 | c3lciA_ |
|
not modelled |
58.1 |
13 |
PDB header:lyase
Chain: A: PDB Molecule:n-acetylneuraminate lyase;
PDBTitle: the d-sialic acid aldolase mutant v251w
|
|
|
| 41 | c3si9B_ |
|
not modelled |
56.4 |
10 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from bartonella2 henselae
|
|
|
| 42 | c3vniC_ |
|
not modelled |
54.3 |
13 |
PDB header:isomerase
Chain: C: PDB Molecule:xylose isomerase domain protein tim barrel;
PDBTitle: crystal structures of d-psicose 3-epimerase from clostridium2 cellulolyticum h10 and its complex with ketohexose sugars
|
|
|
| 43 | d1xkya1 |
|
not modelled |
53.4 |
11 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
|
|
| 44 | c6h4eB_ |
|
not modelled |
52.4 |
15 |
PDB header:lyase
Chain: B: PDB Molecule:putative n-acetylneuraminate lyase;
PDBTitle: proteus mirabilis n-acetylneuraminate lyase
|
|
|
| 45 | c3cprB_ |
|
not modelled |
52.2 |
14 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthetase;
PDBTitle: the crystal structure of corynebacterium glutamicum2 dihydrodipicolinate synthase to 2.2 a resolution
|
|
|
| 46 | c3eb2A_ |
|
not modelled |
51.4 |
16 |
PDB header:lyase
Chain: A: PDB Molecule:putative dihydrodipicolinate synthetase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from2 rhodopseudomonas palustris at 2.0a resolution
|
|
|
| 47 | c1ypxA_ |
|
not modelled |
49.4 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:putative vitamin-b12 independent methionine synthase family
PDBTitle: crystal structure of the putative vitamin-b12 independent methionine2 synthase from listeria monocytogenes, northeast structural genomics3 target lmr13
|
|
|
| 48 | c2a7nA_ |
|
not modelled |
48.8 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:l(+)-mandelate dehydrogenase;
PDBTitle: crystal structure of the g81a mutant of the active chimera of (s)-2 mandelate dehydrogenase
|
|
|
| 49 | c2yxgD_ |
|
not modelled |
48.6 |
11 |
PDB header:lyase
Chain: D: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihyrodipicolinate synthase (dapa)
|
|
|
| 50 | c4i7vD_ |
|
not modelled |
48.4 |
11 |
PDB header:biosynthetic protein
Chain: D: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: agrobacterium tumefaciens dhdps with pyruvate
|
|
|
| 51 | d1p4ca_ |
|
not modelled |
48.4 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
|
|
| 52 | d1hl2a_ |
|
not modelled |
46.9 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
|
|
| 53 | c5k9hA_ |
|
not modelled |
45.6 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:0940_gh29;
PDBTitle: crystal structure of a glycoside hydrolase 29 family member from an2 unknown rumen bacterium
|
|
|
| 54 | c3kwsB_ |
|
not modelled |
44.7 |
16 |
PDB header:isomerase
Chain: B: PDB Molecule:putative sugar isomerase;
PDBTitle: crystal structure of putative sugar isomerase (yp_001305149.1) from2 parabacteroides distasonis atcc 8503 at 1.68 a resolution
|
|
|
| 55 | c3noeA_ |
|
not modelled |
44.0 |
8 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from pseudomonas2 aeruginosa
|
|
|
| 56 | c3lerA_ |
|
not modelled |
43.9 |
16 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from campylobacter2 jejuni subsp. jejuni nctc 11168
|
|
|
| 57 | c2v9dB_ |
|
not modelled |
42.1 |
12 |
PDB header:lyase
Chain: B: PDB Molecule:yage;
PDBTitle: crystal structure of yage, a prophage protein belonging to2 the dihydrodipicolinic acid synthase family from e. coli3 k12
|
|
|
| 58 | d1f74a_ |
|
not modelled |
39.7 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
|
|
| 59 | c3pueA_ |
|
not modelled |
39.3 |
11 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of the complex of dhydrodipicolinate synthase from2 acinetobacter baumannii with lysine at 2.6a resolution
|
|
|
| 60 | c4dppB_ |
|
not modelled |
37.3 |
10 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase 2, chloroplastic;
PDBTitle: the structure of dihydrodipicolinate synthase 2 from arabidopsis2 thaliana
|
|
|
| 61 | c3ilvA_ |
|
not modelled |
36.6 |
14 |
PDB header:ligase
Chain: A: PDB Molecule:glutamine-dependent nad(+) synthetase;
PDBTitle: crystal structure of a glutamine-dependent nad(+) synthetase2 from cytophaga hutchinsonii
|
|
|
| 62 | c2ehhE_ |
|
not modelled |
36.5 |
11 |
PDB header:lyase
Chain: E: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from2 aquifex aeolicus
|
|
|
| 63 | c3s5oA_ |
|
not modelled |
36.1 |
9 |
PDB header:lyase
Chain: A: PDB Molecule:4-hydroxy-2-oxoglutarate aldolase, mitochondrial;
PDBTitle: crystal structure of human 4-hydroxy-2-oxoglutarate aldolase bound to2 pyruvate
|
|
|
| 64 | c4ah7C_ |
|
not modelled |
35.2 |
13 |
PDB header:lyase
Chain: C: PDB Molecule:n-acetylneuraminate lyase;
PDBTitle: structure of wild type stapylococcus aureus n-acetylneuraminic acid2 lyase in complex with pyruvate
|
|
|
| 65 | c3na8A_ |
|
not modelled |
35.0 |
13 |
PDB header:lyase
Chain: A: PDB Molecule:putative dihydrodipicolinate synthetase;
PDBTitle: crystal structure of a putative dihydrodipicolinate synthetase from2 pseudomonas aeruginosa
|
|
|
| 66 | c2wjeA_ |
|
not modelled |
34.4 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:tyrosine-protein phosphatase cpsb;
PDBTitle: crystal structure of the tyrosine phosphatase cps4b from2 steptococcus pneumoniae tigr4.
|
|
|
| 67 | c5afdA_ |
|
not modelled |
32.8 |
15 |
PDB header:lyase
Chain: A: PDB Molecule:n-acetylneuraminate lyase;
PDBTitle: native structure of n-acetylneuramininate lyase (sialic acid aldolase)2 from aliivibrio salmonicida
|
|
|
| 68 | d1r46a2 |
|
not modelled |
32.6 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
|
|
| 69 | c3e96B_ |
|
not modelled |
32.5 |
12 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from bacillus2 clausii
|
|
|
| 70 | c3dz1A_ |
|
not modelled |
32.4 |
12 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from2 rhodopseudomonas palustris at 1.87a resolution
|
|
|
| 71 | c4nq1B_ |
|
not modelled |
32.1 |
9 |
PDB header:lyase
Chain: B: PDB Molecule:4-hydroxy-tetrahydrodipicolinate synthase;
PDBTitle: legionella pneumophila dihydrodipicolinate synthase with first2 substrate pyruvate bound in the active site
|
|
|
| 72 | d1f89a_ |
|
not modelled |
32.0 |
11 |
Fold:Carbon-nitrogen hydrolase
Superfamily:Carbon-nitrogen hydrolase
Family:Nitrilase |
|
|
| 73 | c5c54D_ |
|
not modelled |
31.7 |
13 |
PDB header:lyase
Chain: D: PDB Molecule:dihydrodipicolinate synthase/n-acetylneuraminate lyase;
PDBTitle: crystal structure of a novel n-acetylneuraminic acid lyase from2 corynebacterium glutamicum
|
|
|
| 74 | c4ay8B_ |
|
not modelled |
31.1 |
16 |
PDB header:transferase
Chain: B: PDB Molecule:methylcobalamin\: coenzyme m methyltransferase;
PDBTitle: semet-derivative of a methyltransferase from m. mazei
|
|
|
| 75 | c3lmzA_ |
|
not modelled |
30.8 |
15 |
PDB header:isomerase
Chain: A: PDB Molecule:putative sugar isomerase;
PDBTitle: crystal structure of putative sugar isomerase. (yp_001305105.1) from2 parabacteroides distasonis atcc 8503 at 1.44 a resolution
|
|
|
| 76 | c3bh1A_ |
|
not modelled |
30.7 |
30 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:upf0371 protein dip2346;
PDBTitle: crystal structure of protein dip2346 from corynebacterium diphtheriae
|
|
|
| 77 | c4lsbA_ |
|
not modelled |
29.3 |
7 |
PDB header:lyase
Chain: A: PDB Molecule:lyase/mutase;
PDBTitle: crystal structure of a putative lyase/mutase from burkholderia2 cenocepacia j2315
|
|
|
| 78 | c4zrxA_ |
|
not modelled |
29.3 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:f5/8 type c domain protein;
PDBTitle: crystal structure of a putative alpha-l-fucosidase (bacova_04357) from2 bacteroides ovatus atcc 8483 at 1.59 a resolution
|
|
|
| 79 | c3dcpB_ |
|
not modelled |
28.6 |
37 |
PDB header:hydrolase
Chain: B: PDB Molecule:histidinol-phosphatase;
PDBTitle: crystal structure of the putative histidinol phosphatase hisk from2 listeria monocytogenes. northeast structural genomics consortium3 target lmr141.
|
|
|
| 80 | c4k3zA_ |
|
not modelled |
28.4 |
8 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:d-erythrulose 4-phosphate dehydrogenase;
PDBTitle: crystal structure of d-erythrulose 4-phosphate dehydrogenase from2 brucella melitensis, solved by iodide sad
|
|
|
| 81 | c3fluD_ |
|
not modelled |
27.8 |
8 |
PDB header:lyase
Chain: D: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from the pathogen2 neisseria meningitidis
|
|
|
| 82 | d1hl9a2 |
|
not modelled |
27.6 |
32 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Putative alpha-L-fucosidase, catalytic domain |
|
|
| 83 | c3bi8A_ |
|
not modelled |
27.3 |
9 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: structure of dihydrodipicolinate synthase from clostridium2 botulinum
|
|
|
| 84 | c3wqoB_ |
|
not modelled |
26.7 |
19 |
PDB header:unknown function
Chain: B: PDB Molecule:uncharacterized protein mj1311;
PDBTitle: crystal structure of d-tagatose 3-epimerase-like protein
|
|
|
| 85 | c2qw5B_ |
|
not modelled |
26.6 |
22 |
PDB header:isomerase
Chain: B: PDB Molecule:xylose isomerase-like tim barrel;
PDBTitle: crystal structure of a putative sugar phosphate isomerase/epimerase2 (ava4194) from anabaena variabilis atcc 29413 at 1.78 a resolution
|
|
|
| 86 | c6k0aC_ |
|
not modelled |
26.5 |
15 |
PDB header:rna binding protein/rna
Chain: C: PDB Molecule:ribonuclease p protein component 3;
PDBTitle: cryo-em structure of an archaeal ribonuclease p
|
|
|
| 87 | d2a6na1 |
|
not modelled |
26.1 |
9 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
|
|
| 88 | d1xxxa1 |
|
not modelled |
25.5 |
10 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
|
|
| 89 | c6arhA_ |
|
not modelled |
25.2 |
12 |
PDB header:lyase
Chain: A: PDB Molecule:n-acetylneuraminate lyase;
PDBTitle: crystal structure of human nal at a resolution of 1.6 angstrom
|
|
|
| 90 | c1bplA_ |
|
not modelled |
25.2 |
13 |
PDB header:glycosyltransferase
Chain: A: PDB Molecule:alpha-1,4-glucan-4-glucanohydrolase;
PDBTitle: glycosyltransferase
|
|
|
| 91 | c2yb1A_ |
|
not modelled |
25.0 |
11 |
PDB header:hydrolase
Chain: A: PDB Molecule:amidohydrolase;
PDBTitle: structure of an amidohydrolase from chromobacterium violaceum (efi2 target efi-500202) with bound mn, amp and phosphate.
|
|
|
| 92 | c3h5dD_ |
|
not modelled |
24.7 |
11 |
PDB header:lyase
Chain: D: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: dihydrodipicolinate synthase from drug-resistant streptococcus2 pneumoniae
|
|
|
| 93 | c5ktlA_ |
|
not modelled |
23.9 |
13 |
PDB header:lyase
Chain: A: PDB Molecule:4-hydroxy-tetrahydrodipicolinate synthase;
PDBTitle: dihydrodipicolinate synthase from the industrial and evolutionarily2 important cyanobacteria anabaena variabilis.
|
|
|
| 94 | c3noyA_ |
|
not modelled |
23.8 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase;
PDBTitle: crystal structure of ispg (gcpe)
|
|
|
| 95 | d1goxa_ |
|
not modelled |
23.2 |
9 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
|
|
| 96 | c3fkkA_ |
|
not modelled |
23.2 |
9 |
PDB header:lyase
Chain: A: PDB Molecule:l-2-keto-3-deoxyarabonate dehydratase;
PDBTitle: structure of l-2-keto-3-deoxyarabonate dehydratase
|
|
|
| 97 | c3rpdB_ |
|
not modelled |
22.7 |
11 |
PDB header:transferase
Chain: B: PDB Molecule:methionine synthase (b12-independent);
PDBTitle: the structure of a b12-independent methionine synthase from shewanella2 sp. w3-18-1 in complex with selenomethionine.
|
|
|
| 98 | c4b5nA_ |
|
not modelled |
22.7 |
7 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:oxidoreductase, fmn-binding;
PDBTitle: crystal structure of oxidized shewanella yellow enzyme 4 (sye4)
|
|
|
| 99 | c2hmcA_ |
|
not modelled |
21.8 |
12 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: the crystal structure of dihydrodipicolinate synthase dapa from2 agrobacterium tumefaciens
|
|
|
| 100 | c4a3uB_ |
|
not modelled |
21.5 |
11 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:nadh\:flavin oxidoreductase/nadh oxidase;
PDBTitle: x-structure of the old yellow enzyme homologue from zymomonas mobilis2 (ncr)
|
|
|
| 101 | d1mkza_ |
|
not modelled |
20.9 |
16 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MogA-like |
|
|
| 102 | c6mqhA_ |
|
not modelled |
20.6 |
11 |
PDB header:lyase
Chain: A: PDB Molecule:4-hydroxy-tetrahydrodipicolinate synthase;
PDBTitle: crystal structure of 4-hydroxy-tetrahydrodipicolinate synthase (htpa2 synthase) from burkholderia mallei
|
|
|
| 103 | d2f6ua1 |
|
not modelled |
20.5 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
|
|
| 104 | c4icnB_ |
|
not modelled |
20.3 |
11 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: dihydrodipicolinate synthase from shewanella benthica
|
|
|