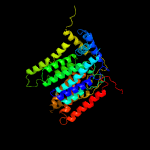
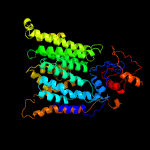
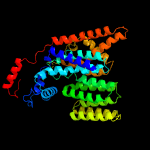
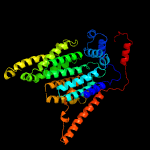
1 c6p2rB_
99.7
16
PDB header: transferaseChain: B: PDB Molecule: dolichyl-phosphate-mannose--protein mannosyltransferase 2;PDBTitle: structure of s. cerevisiae protein o-mannosyltransferase pmt1-pmt22 complex bound to the sugar donor
2 c5f15A_
99.7
13
PDB header: transferaseChain: A: PDB Molecule: 4-amino-4-deoxy-l-arabinose (l-ara4n) transferase;PDBTitle: crystal structure of arnt from cupriavidus metallidurans bound to2 undecaprenyl phosphate
3 c6p25A_
99.6
13
PDB header: transferaseChain: A: PDB Molecule: dolichyl-phosphate-mannose--protein mannosyltransferase 1;PDBTitle: structure of s. cerevisiae protein o-mannosyltransferase pmt1-pmt22 complex bound to the sugar donor and a peptide acceptor
4 c3wajA_
99.3
12
PDB header: transferaseChain: A: PDB Molecule: transmembrane oligosaccharyl transferase;PDBTitle: crystal structure of the archaeoglobus fulgidus2 oligosaccharyltransferase (o29867_arcfu) complex with zn and sulfate
5 c3rceA_
99.1
10
PDB header: transferase/peptideChain: A: PDB Molecule: oligosaccharide transferase to n-glycosylate proteins;PDBTitle: bacterial oligosaccharyltransferase pglb
6 c6eznF_
98.8
13
PDB header: membrane proteinChain: F: PDB Molecule: dolichyl-diphosphooligosaccharide--proteinPDBTitle: cryo-em structure of the yeast oligosaccharyltransferase (ost) complex
7 d1lura_
56.4
17
Fold: SupersandwichSuperfamily: Galactose mutarotase-likeFamily: Aldose 1-epimerase (mutarotase)
8 d1wxma1
50.2
25
Fold: beta-Grasp (ubiquitin-like)Superfamily: Ubiquitin-likeFamily: Ras-binding domain, RBD
9 c4rdbA_
45.8
27
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: immunoreactive 32 kda antigen pg49;PDBTitle: crystal structure of an immunoreactive 32 kda antigen pg49 (pg_0181)2 from porphyromonas gingivalis w83 at 1.45 a resolution (psi community3 target, nakayama)
10 c5n1tM_
45.2
21
PDB header: oxidoreductaseChain: M: PDB Molecule: copc;PDBTitle: crystal structure of complex between flavocytochrome c and copper2 chaperone copc from t. paradoxus
11 c4adjB_
43.6
18
PDB header: viral proteinChain: B: PDB Molecule: e1 envelope glycoprotein;PDBTitle: crystal structure of the rubella virus glycoprotein e1 in its2 post-fusion form crystallized in presence of 1mm of calcium acetate
12 c6cd2C_
37.9
14
PDB header: membrane protein/chaperoneChain: C: PDB Molecule: outer membrane usher protein papc;PDBTitle: crystal structure of the papc usher bound to the chaperone-adhesin2 papd-papg
13 c6nfqC_
31.5
13
PDB header: metal binding proteinChain: C: PDB Molecule: copc;PDBTitle: copc from pseudomonas fluorescens
14 c3ohnA_
30.2
15
PDB header: membrane proteinChain: A: PDB Molecule: outer membrane usher protein fimd;PDBTitle: crystal structure of the fimd translocation domain
15 d2f2ha1
29.2
28
Fold: Putative glucosidase YicI, C-terminal domainSuperfamily: Putative glucosidase YicI, C-terminal domainFamily: Putative glucosidase YicI, C-terminal domain
16 c4dipD_
24.5
18
PDB header: isomeraseChain: D: PDB Molecule: peptidyl-prolyl cis-trans isomerase fkbp14;PDBTitle: crystal structure of human peptidyl-prolyl cis-trans isomerase fkbp14
17 c2w1wB_
22.1
13
PDB header: hydrolaseChain: B: PDB Molecule: lipolytic enzyme, g-d-s-l;PDBTitle: native structure of a family 35 carbohydrate binding module2 from clostridium thermocellum
18 c5yxiA_
21.3
23
PDB header: de novo proteinChain: A: PDB Molecule: drafx6;PDBTitle: designed protein drafx6
19 c3k8rA_
19.7
17
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of protein of unknown function (yp_427503.1) from2 rhodospirillum rubrum atcc 11170 at 2.75 a resolution
20 c4jjaA_
19.1
12
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein;PDBTitle: crystal structure of a duf1343 family protein (bf0379) from2 bacteroides fragilis nctc 9343 at 1.30 a resolution
21 c2yevB_
not modelled
18.9
17
PDB header: electron transportChain: B: PDB Molecule: cytochrome c oxidase subunit 2;PDBTitle: structure of caa3-type cytochrome oxidase
22 c6gszA_
not modelled
17.6
12
PDB header: hydrolaseChain: A: PDB Molecule: alpha-l-rhamnosidase;PDBTitle: crystal structure of native alfa-l-rhamnosidase from aspergillus2 terreus
23 c2kzbA_
not modelled
17.4
10
PDB header: protein transportChain: A: PDB Molecule: autophagy-related protein 19;PDBTitle: solution structure of alpha-mannosidase binding domain of atg19
24 c4mspB_
not modelled
16.8
18
PDB header: isomeraseChain: B: PDB Molecule: peptidyl-prolyl cis-trans isomerase fkbp14;PDBTitle: crystal structure of human peptidyl-prolyl cis-trans isomerase fkbp222 (aka fkbp14) containing two ef-hand motifs
25 d1nsza_
not modelled
16.7
17
Fold: SupersandwichSuperfamily: Galactose mutarotase-likeFamily: Aldose 1-epimerase (mutarotase)
26 c2okxB_
not modelled
16.5
18
PDB header: hydrolaseChain: B: PDB Molecule: rhamnosidase b;PDBTitle: crystal structure of gh78 family rhamnosidase of bacillus sp. gl1 at2 1.9 a
27 d1ulva2
not modelled
16.2
24
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: E-set domains of sugar-utilizing enzymes
28 c2vzqA_
not modelled
16.1
16
PDB header: hydrolaseChain: A: PDB Molecule: exo-beta-d-glucosaminidase;PDBTitle: c-terminal cbm35 from amycolatopsis orientalis exo-chitosanase csxa in2 complex with digalacturonic acid
29 c5ir6B_
not modelled
15.9
7
PDB header: oxidoreductaseChain: B: PDB Molecule: bd-type quinol oxidase subunit ii;PDBTitle: the structure of bd oxidase from geobacillus thermodenitrificans
30 c3mwxA_
not modelled
15.9
12
PDB header: isomeraseChain: A: PDB Molecule: aldose 1-epimerase;PDBTitle: crystal structure of a putative galactose mutarotase (bsu18360) from2 bacillus subtilis at 1.45 a resolution
31 d1j0ha2
not modelled
15.6
20
Fold: Glycosyl hydrolase domainSuperfamily: Glycosyl hydrolase domainFamily: alpha-Amylases, C-terminal beta-sheet domain
32 d2o34a1
not modelled
15.4
23
Fold: T-foldSuperfamily: ApbE-likeFamily: DVU1097-like
33 c3nreB_
not modelled
14.9
13
PDB header: isomeraseChain: B: PDB Molecule: aldose 1-epimerase;PDBTitle: crystal structure of a putative aldose 1-epimerase (b2544) from2 escherichia coli k12 at 1.59 a resolution
34 d1ix2a_
not modelled
14.8
18
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: Copper resistance protein C (CopC, PcoC)
35 c4dtfA_
not modelled
14.7
16
PDB header: toxinChain: A: PDB Molecule: vgrg protein;PDBTitle: structure of a vgrg vibrio cholerae toxin acd domain in complex with2 amp-pnp and mg++
36 c4gxbA_
not modelled
14.6
25
PDB header: protein transport/cell adhesionChain: A: PDB Molecule: sorting nexin-17;PDBTitle: structure of the snx17 atypical ferm domain bound to the npxy motif of2 p-selectin
37 c3i31A_
not modelled
14.6
19
PDB header: rna binding protein,hydrolaseChain: A: PDB Molecule: heat resistant rna dependent atpase;PDBTitle: hera helicase rna binding domain is an rrm fold
38 c3dcdA_
not modelled
14.6
13
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: galactose mutarotase related enzyme;PDBTitle: x-ray structure of the galactose mutarotase related enzyme q5fkd7 from2 lactobacillus acidophilus at the resolution 1.9a. northeast3 structural genomics consortium target lar33.
39 c2bgpA_
not modelled
14.6
11
PDB header: carbohydrate binding proteinChain: A: PDB Molecule: endo-b1,4-mannanase 5c;PDBTitle: mannan binding module from man5c in bound conformation
40 c5w21B_
not modelled
14.2
21
PDB header: hydrolase/protein bindingChain: B: PDB Molecule: fibroblast growth factor 23;PDBTitle: crystal structure of a 1:1:1 fgf23-fgfr1c-aklotho ternary complex
41 c2w3jA_
not modelled
14.2
16
PDB header: sugar-binding proteinChain: A: PDB Molecule: carbohydrate binding module;PDBTitle: structure of a family 35 carbohydrate binding module from2 an environmental isolate
42 c4k05B_
not modelled
14.1
12
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: conserved hypothetical exported protein;PDBTitle: crystal structure of a duf1343 family protein (bf0371) from2 bacteroides fragilis nctc 9343 at 1.65 a resolution
43 c3rfuC_
not modelled
13.7
18
PDB header: hydrolase, membrane proteinChain: C: PDB Molecule: copper efflux atpase;PDBTitle: crystal structure of a copper-transporting pib-type atpase
44 c6e14D_
not modelled
13.7
13
PDB header: membrane proteinChain: D: PDB Molecule: fimbrial biogenesis outer membrane usher protein;PDBTitle: handover mechanism of the growing pilus by the bacterial outer2 membrane usher fimd
45 d1c3ga2
not modelled
13.4
19
Fold: HSP40/DnaJ peptide-binding domainSuperfamily: HSP40/DnaJ peptide-binding domainFamily: HSP40/DnaJ peptide-binding domain
46 c6e8aA_
not modelled
13.3
19
PDB header: unknown functionChain: A: PDB Molecule: duf1795 domain-containing protein;PDBTitle: crystal structure of dcrb from salmonella enterica at 1.92 angstroms2 resolution
47 c4rnlB_
not modelled
12.9
23
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: possible galactose mutarotase;PDBTitle: the crystal structure of a possible galactose mutarotase from2 streptomyces platensis subsp. rosaceus
48 d1od3a_
not modelled
12.7
19
Fold: Galactose-binding domain-likeSuperfamily: Galactose-binding domain-likeFamily: Family 6 carbohydrate binding module, CBM6
49 d1i5pa1
not modelled
12.7
19
Fold: Galactose-binding domain-likeSuperfamily: Galactose-binding domain-likeFamily: delta-Endotoxin, C-terminal domain
50 c2hbpA_
not modelled
12.2
18
PDB header: endocytosis, protein bindingChain: A: PDB Molecule: cytoskeleton assembly control protein sla1;PDBTitle: solution structure of sla1 homology domain 1
51 c3oisA_
not modelled
12.2
6
PDB header: hydrolaseChain: A: PDB Molecule: cysteine protease;PDBTitle: crystal structure xylellain, a cysteine protease from xylella2 fastidiosa
52 c5xyiD_
not modelled
12.0
21
PDB header: ribosomeChain: D: PDB Molecule: ribosomal protein s3, putative;PDBTitle: small subunit of trichomonas vaginalis ribosome
53 d2i6va1
not modelled
11.9
26
Fold: PDZ domain-likeSuperfamily: PDZ domain-likeFamily: EpsC C-terminal domain-like
54 c2auwB_
not modelled
11.8
16
PDB header: unknown functionChain: B: PDB Molecule: hypothetical protein ne0471;PDBTitle: crystal structure of putative dna binding protein ne0471 from2 nitrosomonas europaea atcc 19718
55 c1xrdA_
not modelled
11.7
30
PDB header: membrane proteinChain: A: PDB Molecule: light-harvesting protein b-880, alpha chain;PDBTitle: light-harvesting complex 1 alfa subunit from wild-type2 rhodospirillum rubrum
56 d1xrda1
not modelled
11.7
30
Fold: Light-harvesting complex subunitsSuperfamily: Light-harvesting complex subunitsFamily: Light-harvesting complex subunits
57 c5awqA_
not modelled
11.6
10
PDB header: hydrolaseChain: A: PDB Molecule: isomaltodextranase;PDBTitle: arthrobacter globiformis t6 isomalto-dextranse complexed with panose
58 c5hj1A_
not modelled
11.6
25
PDB header: hydrolaseChain: A: PDB Molecule: pullulanase c protein;PDBTitle: crystal structure of pdz domain of pullulanase c protein of type ii2 secretion system from klebsiella pneumoniae in complex with fatty3 acid
59 c6et5b_
not modelled
11.6
27
PDB header: photosynthesisChain: B: PDB Molecule: PDBTitle: reaction centre light harvesting complex 1 from blc. virids
60 c3imhB_
not modelled
11.4
14
PDB header: isomeraseChain: B: PDB Molecule: galactose-1-epimerase;PDBTitle: crystal structure of galactose 1-epimerase from lactobacillus2 acidophilus ncfm
61 c3n6yA_
not modelled
11.4
13
PDB header: unknown functionChain: A: PDB Molecule: immunoglobulin-like protein;PDBTitle: crystal structure of an immunoglobulin-like protein (pa1606) from2 pseudomonas aeruginosa at 1.50 a resolution
62 c1sr9A_
not modelled
11.3
13
PDB header: transferaseChain: A: PDB Molecule: 2-isopropylmalate synthase;PDBTitle: crystal structure of leua from mycobacterium tuberculosis
63 d2c9qa1
not modelled
11.3
21
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: Copper resistance protein C (CopC, PcoC)
64 d2azeb1
not modelled
11.3
3
Fold: E2F-DP heterodimerization regionSuperfamily: E2F-DP heterodimerization regionFamily: E2F dimerization segment
65 c2xznC_
not modelled
11.1
17
PDB header: ribosomeChain: C: PDB Molecule: kh domain containing protein;PDBTitle: crystal structure of the eukaryotic 40s ribosomal2 subunit in complex with initiation factor 1. this file3 contains the 40s subunit and initiation factor for4 molecule 2
66 c2vqiA_
not modelled
11.1
6
PDB header: transportChain: A: PDB Molecule: outer membrane usher protein papc;PDBTitle: structure of the p pilus usher (papc) translocation pore
67 c4hs5B_
not modelled
10.8
12
PDB header: metal binding proteinChain: B: PDB Molecule: protein cyay;PDBTitle: frataxin from psychromonas ingrahamii as a model to study stability2 modulation within cyay protein family
68 d1dp4a_
not modelled
10.6
27
Fold: Periplasmic binding protein-like ISuperfamily: Periplasmic binding protein-like IFamily: L-arabinose binding protein-like
69 d1t3qa2
not modelled
10.6
18
Fold: beta-Grasp (ubiquitin-like)Superfamily: 2Fe-2S ferredoxin-likeFamily: 2Fe-2S ferredoxin domains from multidomain proteins
70 c4ifdE_
not modelled
10.5
27
PDB header: hydrolase/rnaChain: E: PDB Molecule: exosome complex component rrp42;PDBTitle: crystal structure of an 11-subunit eukaryotic exosome complex bound to2 rna
71 c3i38E_
not modelled
10.5
19
PDB header: chaperoneChain: E: PDB Molecule: putative chaperone dnaj;PDBTitle: structure of a putative chaperone protein dnaj from klebsiella2 pneumoniae subsp. pneumoniae mgh 78578
72 c3i38C_
not modelled
10.5
19
PDB header: chaperoneChain: C: PDB Molecule: putative chaperone dnaj;PDBTitle: structure of a putative chaperone protein dnaj from klebsiella2 pneumoniae subsp. pneumoniae mgh 78578
73 c4v1ao_
not modelled
10.4
10
PDB header: ribosomeChain: O: PDB Molecule: PDBTitle: structure of the large subunit of the mammalian mitoribosome, part 22 of 2
74 c2wzpR_
not modelled
10.3
16
PDB header: viral proteinChain: R: PDB Molecule: lactococcal phage p2 orf16;PDBTitle: structures of lactococcal phage p2 baseplate shed light on2 a novel mechanism of host attachment and activation in3 siphoviridae
75 c2kijA_
not modelled
10.3
14
PDB header: hydrolaseChain: A: PDB Molecule: copper-transporting atpase 1;PDBTitle: solution structure of the actuator domain of the copper-2 transporting atpase atp7a
76 c4jpdA_
not modelled
10.3
18
PDB header: metal binding proteinChain: A: PDB Molecule: protein cyay;PDBTitle: the structure of cyay from burkholderia cenocepacia
77 c1fftG_
not modelled
10.2
16
PDB header: oxidoreductaseChain: G: PDB Molecule: ubiquinol oxidase;PDBTitle: the structure of ubiquinol oxidase from escherichia coli
78 c2a8lB_
not modelled
10.2
12
PDB header: hydrolaseChain: B: PDB Molecule: threonine aspartase 1;PDBTitle: crystal structure of human taspase1 (t234a mutant)
79 c3ccjU_
not modelled
10.2
25
PDB header: ribosomeChain: U: PDB Molecule: 50s ribosomal protein l24e;PDBTitle: structure of anisomycin resistant 50s ribosomal subunit: 23s rrna2 mutation c2534u
80 d1vqou1
not modelled
10.2
25
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: Ribosomal protein L24e
81 c6adqW_
not modelled
10.2
20
PDB header: electron transportChain: W: PDB Molecule: lpqe protein;PDBTitle: respiratory complex ciii2civ2sod2 from mycobacterium smegmatis
82 c2zkru_
not modelled
10.1
44
PDB header: ribosomal protein/rnaChain: U: PDB Molecule: rna expansion segment es41;PDBTitle: structure of a mammalian ribosomal 60s subunit within an 80s complex2 obtained by docking homology models of the rna and proteins into an3 8.7 a cryo-em map
83 c3zeyX_
not modelled
10.1
17
PDB header: ribosomeChain: X: PDB Molecule: 40s ribosomal protein s3, putative;PDBTitle: high-resolution cryo-electron microscopy structure of the trypanosoma2 brucei ribosome
84 c3ggqA_
not modelled
10.1
18
PDB header: viral proteinChain: A: PDB Molecule: capsid protein;PDBTitle: dimerization of hepatitis e virus capsid protein e2s domain is2 essential for virus-host interaction
85 d1wf9a1
not modelled
10.1
18
Fold: beta-Grasp (ubiquitin-like)Superfamily: Ubiquitin-likeFamily: Ubiquitin-related
86 c3fljA_
not modelled
10.0
23
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein conserved in bacteria with aPDBTitle: crystal structure of uncharacterized protein conserved in bacteria2 with a cystatin-like fold (yp_168589.1) from silicibacter pomeroyi3 dss-3 at 2.00 a resolution
87 d1kn0a_
not modelled
10.0
13
Fold: dsRBD-likeSuperfamily: dsRNA-binding domain-likeFamily: The homologous-pairing domain of Rad52 recombinase
88 c2x8nA_
not modelled
10.0
13
PDB header: structural genomicsChain: A: PDB Molecule: cv0863;PDBTitle: solution nmr structure of uncharacterized protein cv0863 from2 chromobacterium violaceum. northeast structural genomics target3 (nesg) target cvt3. ocsp target cv0863.
89 c3wmmY_
not modelled
10.0
28
PDB header: photosynthesisChain: Y: PDB Molecule: lh1 alpha polypeptide;PDBTitle: crystal structure of the lh1-rc complex from thermochromatium tepidum2 in c2 form
90 c1hyoB_
not modelled
9.9
19
PDB header: hydrolaseChain: B: PDB Molecule: fumarylacetoacetate hydrolase;PDBTitle: crystal structure of fumarylacetoacetate hydrolase2 complexed with 4-(hydroxymethylphosphinoyl)-3-oxo-butanoic3 acid
91 d1hdgo2
not modelled
9.9
42
Fold: FwdE/GAPDH domain-likeSuperfamily: Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domainFamily: GAPDH-like
92 d1ew4a_
not modelled
9.9
15
Fold: N domain of copper amine oxidase-likeSuperfamily: Frataxin/Nqo15-likeFamily: Frataxin-like
93 c3o1hB_
not modelled
9.9
12
PDB header: signaling proteinChain: B: PDB Molecule: periplasmic protein tort;PDBTitle: crystal structure of the tors sensor domain - tort complex in the2 presence of tmao
94 d1uy4a_
not modelled
9.9
12
Fold: Galactose-binding domain-likeSuperfamily: Galactose-binding domain-likeFamily: Family 6 carbohydrate binding module, CBM6
95 c1s1iS_
not modelled
9.8
19
PDB header: ribosomeChain: S: PDB Molecule: 60s ribosomal protein l24-a;PDBTitle: structure of the ribosomal 80s-eef2-sordarin complex from yeast2 obtained by docking atomic models for rna and protein components into3 a 11.7 a cryo-em map. this file, 1s1i, contains 60s subunit. the 40s4 ribosomal subunit is in file 1s1h.
96 d1pjxa_
not modelled
9.8
5
Fold: 6-bladed beta-propellerSuperfamily: Calcium-dependent phosphotriesteraseFamily: SGL-like
97 c1s1hC_
not modelled
9.8
21
PDB header: ribosomeChain: C: PDB Molecule: 40s ribosomal protein s3;PDBTitle: structure of the ribosomal 80s-eef2-sordarin complex from yeast2 obtained by docking atomic models for rna and protein components into3 a 11.7 a cryo-em map. this file, 1s1h, contains 40s subunit. the 60s4 ribosomal subunit is in file 1s1i.
98 c3izbB_
not modelled
9.8
21
PDB header: ribosomeChain: B: PDB Molecule: 40s ribosomal protein rps3 (s3p);PDBTitle: localization of the small subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
99 c4go6C_
not modelled
9.8
35
PDB header: protein bindingChain: C: PDB Molecule: hcf n-terminal chain 1;PDBTitle: crystal structure of hcf-1 self-association sequence 1