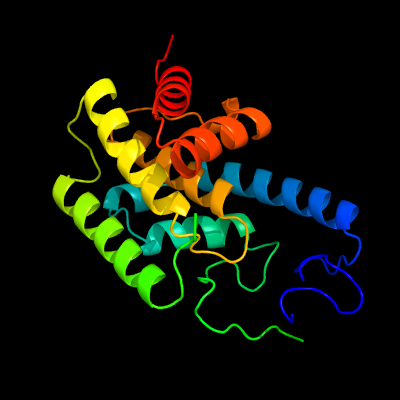
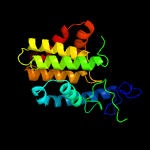
1 c2vzaD_
100.0
27
PDB header: cell adhesionChain: D: PDB Molecule: cell filamentation protein;PDBTitle: type iv secretion system effector protein bepa
2 c4npsA_
100.0
25
PDB header: cell adhesionChain: A: PDB Molecule: bartonella effector protein (bep) substrate of virb t4ss;PDBTitle: crystal structure of bep1 protein (virb-translocated bartonella2 effector protein) from bartonella clarridgeiae
3 c4m16A_
100.0
26
PDB header: cell adhesionChain: A: PDB Molecule: bartonella effector protein (bep) substrate of virb t4ss;PDBTitle: crystal structure of the n-terminal fic domain of bartonella effector2 protein (bep); substrate of virb t4ss (virb-translocated bep effector3 protein) from bartonella sp. ar 15-3
4 c4lu4A_
100.0
26
PDB header: transferaseChain: A: PDB Molecule: putative cell filamentation protein;PDBTitle: crystal structure of the n-terminal fic domain of a putative cell2 filamentation protein (virb-translocated bep effector protein) from3 bartonella quintana
5 c4xi8B_
100.0
22
PDB header: protein bindingChain: B: PDB Molecule: bartonella effector protein (bep) substrate of virb t4ss;PDBTitle: crystal structure of the fic domain of bep5 protein (virb-translocated2 bartonella effector protein) from bartonella clarridgeiae
6 c4py3B_
100.0
23
PDB header: protein bindingChain: B: PDB Molecule: bartonella effector protein (bep) substrate of virb t4ss;PDBTitle: crystal structure of the n-terminal fic domain of bep8 protein (virb-2 translocated bartonella effector protein) from bartonella sp. 1-1c
7 c3shgA_
100.0
28
PDB header: transferase/protein bindingChain: A: PDB Molecule: vbht;PDBTitle: vbht fic protein from bartonella schoenbuchensis in complex with vbha2 antitoxin
8 c5jffC_
100.0
30
PDB header: transferaseChain: C: PDB Molecule: probable adenosine monophosphate-protein transferase fic;PDBTitle: e. coli ecfict mutant g55r in complex with ecfica
9 c5nwfB_
100.0
27
PDB header: toxinChain: B: PDB Molecule: fic family protein;PDBTitle: enterococcus faecalis fic protein (h111a).
10 c2f6sA_
100.0
26
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: cell filamentation protein, putative;PDBTitle: structure of cell filamentation protein (fic) from helicobacter pylori
11 d2f6sa1
100.0
25
Fold: Fic-likeSuperfamily: Fic-likeFamily: Fic-like
12 d2g03a1
100.0
25
Fold: Fic-likeSuperfamily: Fic-likeFamily: Fic-like
13 c4u0zH_
100.0
17
PDB header: transferaseChain: H: PDB Molecule: adenosine monophosphate-protein transferase ficd;PDBTitle: eukaryotic fic domain containing protein with bound apcpp
14 c3cucB_
99.9
14
PDB header: signaling proteinChain: B: PDB Molecule: protein of unknown function with a fic domain;PDBTitle: crystal structure of a fic domain containing signaling protein2 (bt_2513) from bacteroides thetaiotaomicron vpi-5482 at 2.71 a3 resolution
15 c4rglA_
99.9
20
PDB header: dna binding proteinChain: A: PDB Molecule: filamentation induced by camp protein fic;PDBTitle: crystal structure of a fic family protein (dde_2494) from2 desulfovibrio desulfuricans g20 at 2.70 a resolution
16 c5jj6A_
99.9
17
PDB header: transferaseChain: A: PDB Molecule: adenosine monophosphate-protein transferase ficd homolog;PDBTitle: fic-1 (aa134 - 508) from c. elegans
17 c3eqxB_
99.9
13
PDB header: dna binding proteinChain: B: PDB Molecule: fic domain containing transcriptional regulator;PDBTitle: crystal structure of a fic family protein (so_4266) from shewanella2 oneidensis at 1.6 a resolution
18 c4x2eA_
99.9
16
PDB header: transferaseChain: A: PDB Molecule: fic family protein putative filamentation induced by campPDBTitle: clostridium difficile wild type fic protein
19 c3n3vA_
99.9
17
PDB header: transferaseChain: A: PDB Molecule: adenosine monophosphate-protein transferase ibpa;PDBTitle: crystal structure of ibpafic2-h3717a in complex with adenylylated2 cdc42
20 c3dd7A_
99.0
16
PDB header: ribosome inhibitorChain: A: PDB Molecule: death on curing protein;PDBTitle: structure of doch66y in complex with the c-terminal domain of phd
21 c3letB_
not modelled
98.3
17
PDB header: transferaseChain: B: PDB Molecule: adenosine monophosphate-protein transferase vops;PDBTitle: crystal structure of fic domain containing ampylator, vops
22 d1s0pa_
not modelled
42.4
13
Fold: N-terminal domain of adenylylcyclase associated protein, CAPSuperfamily: N-terminal domain of adenylylcyclase associated protein, CAPFamily: N-terminal domain of adenylylcyclase associated protein, CAP
23 c3n0aA_
not modelled
40.3
10
PDB header: hydrolaseChain: A: PDB Molecule: tyrosine-protein phosphatase auxilin;PDBTitle: crystal structure of auxilin (40-400)
24 d3etja2
not modelled
40.3
39
Fold: PreATP-grasp domainSuperfamily: PreATP-grasp domainFamily: BC N-terminal domain-like
25 c2c46B_
not modelled
37.1
9
PDB header: transferaseChain: B: PDB Molecule: mrna capping enzyme;PDBTitle: crystal structure of the human rna guanylyltransferase and 5'-2 phosphatase
26 c3awfC_
not modelled
34.4
14
PDB header: hydrolase, membrane proteinChain: C: PDB Molecule: voltage-sensor containing phosphatase;PDBTitle: crystal structure of pten-like domain of ci-vsp (236-576)
27 c2imgA_
not modelled
30.6
29
PDB header: hydrolaseChain: A: PDB Molecule: dual specificity protein phosphatase 23;PDBTitle: crystal structure of dual specificity protein phosphatase2 23 from homo sapiens in complex with ligand malate ion
28 c3rgqA_
not modelled
29.3
18
PDB header: hydrolaseChain: A: PDB Molecule: protein-tyrosine phosphatase mitochondrial 1;PDBTitle: crystal structure of ptpmt1 in complex with pi(5)p
29 d1yl7a1
not modelled
28.7
14
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain
30 c2ee7A_
not modelled
28.5
7
PDB header: structural proteinChain: A: PDB Molecule: sperm flagellar protein 1;PDBTitle: solution structure of the ch domain from human sperm2 flagellar protein 1
31 c1yn9B_
not modelled
28.3
17
PDB header: hydrolaseChain: B: PDB Molecule: polynucleotide 5'-phosphatase;PDBTitle: crystal structure of baculovirus rna 5'-phosphatase2 complexed with phosphate
32 d1ohea2
not modelled
27.2
13
Fold: (Phosphotyrosine protein) phosphatases IISuperfamily: (Phosphotyrosine protein) phosphatases IIFamily: Dual specificity phosphatase-like
33 c1d5rA_
not modelled
25.2
17
PDB header: hydrolaseChain: A: PDB Molecule: phosphoinositide phosphatase pten;PDBTitle: crystal structure of the pten tumor suppressor
34 d1mkpa_
not modelled
24.8
17
Fold: (Phosphotyrosine protein) phosphatases IISuperfamily: (Phosphotyrosine protein) phosphatases IIFamily: Dual specificity phosphatase-like
35 c4nyhB_
not modelled
24.5
21
PDB header: hydrolaseChain: B: PDB Molecule: rna/rnp complex-1-interacting phosphatase;PDBTitle: orthorhombic crystal form of pir1 dual specificity phosphatase core
36 c2lcyA_
not modelled
24.0
41
PDB header: viral proteinChain: A: PDB Molecule: virion spike glycoprotein;PDBTitle: nmr structure of the complete internal fusion loop from ebolavirus gp22 at ph 5.5
37 c6i28A_
not modelled
22.7
15
PDB header: viral proteinChain: A: PDB Molecule: orf98 ptp-2;PDBTitle: crystal structure of cydia pomonella ptp-2 phosphatase
38 c3s4eA_
not modelled
22.3
13
PDB header: hydrolaseChain: A: PDB Molecule: dual specificity protein phosphatase 19;PDBTitle: crystal structrue of a novel mitogen-activated protein kinase2 phosphatase, skrp1
39 c2htfA_
not modelled
22.0
35
PDB header: transferaseChain: A: PDB Molecule: dna polymerase mu;PDBTitle: the solution structure of the brct domain from human2 polymerase reveals homology with the tdt brct domain
40 c1zzwA_
not modelled
21.7
17
PDB header: hydrolaseChain: A: PDB Molecule: dual specificity protein phosphatase 10;PDBTitle: crystal structure of catalytic domain of human map kinase2 phosphatase 5
41 c2wgpA_
not modelled
20.8
22
PDB header: hydrolaseChain: A: PDB Molecule: dual specificity protein phosphatase 14;PDBTitle: crystal structure of human dual specificity phosphatase 14
42 c2y96A_
not modelled
20.2
21
PDB header: hydrolaseChain: A: PDB Molecule: dual specificity phosphatase dupd1;PDBTitle: structure of human dual-specificity phosphatase 27
43 c2gwoC_
not modelled
19.8
21
PDB header: hydrolaseChain: C: PDB Molecule: dual specificity protein phosphatase 13;PDBTitle: crystal structure of tmdp
44 c2i6oA_
not modelled
19.3
23
PDB header: hydrolaseChain: A: PDB Molecule: sulfolobus solfataricus protein tyrosinePDBTitle: crystal structure of the complex of the archaeal sulfolobus2 ptp-fold phosphatase with phosphopeptides n-g-(p)y-k-n
45 c2nt2C_
not modelled
19.0
29
PDB header: hydrolaseChain: C: PDB Molecule: protein phosphatase slingshot homolog 2;PDBTitle: crystal structure of slingshot phosphatase 2
46 c2e0tA_
not modelled
18.4
22
PDB header: hydrolaseChain: A: PDB Molecule: dual specificity phosphatase 26;PDBTitle: crystal structure of catalytic domain of dual specificity phosphatase2 26, ms0830 from homo sapiens
47 d1s2xa_
not modelled
18.1
21
Fold: STAT-likeSuperfamily: Cag-ZFamily: Cag-Z
48 c1s2xA_
not modelled
18.1
21
PDB header: unknown functionChain: A: PDB Molecule: cag-z;PDBTitle: crystal structure of cag-z from helicobacter pylori
49 c3emuA_
not modelled
17.9
9
PDB header: hydrolaseChain: A: PDB Molecule: leucine rich repeat and phosphatase domain containingPDBTitle: crystal structure of a leucine rich repeat and phosphatase domain2 containing protein from entamoeba histolytica
50 c1wrmA_
not modelled
17.9
17
PDB header: hydrolaseChain: A: PDB Molecule: dual specificity phosphatase 22;PDBTitle: crystal structure of jsp-1
51 c1yl7F_
not modelled
17.8
14
PDB header: oxidoreductaseChain: F: PDB Molecule: dihydrodipicolinate reductase;PDBTitle: the crystal structure of mycobacterium tuberculosis2 dihydrodipicolinate reductase (rv2773c) in complex with nadh (crystal3 form c)
52 d1diha1
not modelled
17.7
33
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain
53 c4ki9A_
not modelled
17.6
17
PDB header: hydrolaseChain: A: PDB Molecule: dual specificity protein phosphatase 12;PDBTitle: crystal structure of the catalytic domain of human dusp12 at 2.0 a2 resolution
54 c5jnmA_
not modelled
17.6
21
PDB header: oxidoreductaseChain: A: PDB Molecule: mannitol-1-phosphate 5-dehydrogenase;PDBTitle: crystal structure of mtld from staphylococcus aureus at 1.7-angstrom2 resolution
55 c2r0bA_
not modelled
17.5
17
PDB header: hydrolaseChain: A: PDB Molecule: serine/threonine/tyrosine-interacting protein;PDBTitle: crystal structure of human tyrosine phosphatase-like2 serine/threonine/tyrosine-interacting protein
56 c2m3vA_
not modelled
16.8
18
PDB header: hydrolaseChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: solution structure of tyrosine phosphatase related to biofilm2 formation a (tpba) from pseudomonas aeruginosa
57 c2oudA_
not modelled
16.1
17
PDB header: hydrolaseChain: A: PDB Molecule: dual specificity protein phosphatase 10;PDBTitle: crystal structure of the catalytic domain of human mkp5
58 c5kt0A_
not modelled
15.8
24
PDB header: oxidoreductaseChain: A: PDB Molecule: 4-hydroxy-tetrahydrodipicolinate reductase;PDBTitle: dihydrodipicolinate reductase from the industrial and evolutionarily2 important cyanobacteria anabaena variabilis.
59 c4kyrA_
not modelled
15.4
18
PDB header: hydrolase, sugar binding proteinChain: A: PDB Molecule: phosphoglucan phosphatase lsf2, chloroplastic;PDBTitle: structure of a product bound plant phosphatase
60 c3ijpA_
not modelled
15.3
33
PDB header: oxidoreductaseChain: A: PDB Molecule: dihydrodipicolinate reductase;PDBTitle: crystal structure of dihydrodipicolinate reductase from bartonella2 henselae at 2.0a resolution
61 d1u8fo1
not modelled
14.9
18
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain
62 c4r30C_
not modelled
14.8
33
PDB header: hydrolaseChain: C: PDB Molecule: laforin;PDBTitle: structure of human laforin dual specificity phosphatase domain
63 c2g6zB_
not modelled
14.7
22
PDB header: hydrolaseChain: B: PDB Molecule: dual specificity protein phosphatase 5;PDBTitle: crystal structure of human dusp5
64 c4iqzD_
not modelled
14.4
31
PDB header: unknown functionChain: D: PDB Molecule: dna-directed rna polymerase subunit beta';PDBTitle: the crystal structure of a large insert in rna polymerase (rpoc)2 subunit from e. coli
65 d1vm6a3
not modelled
14.4
40
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain
66 d1j0xo1
not modelled
14.3
18
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain
67 d1i9sa_
not modelled
14.2
13
Fold: (Phosphotyrosine protein) phosphatases IISuperfamily: (Phosphotyrosine protein) phosphatases IIFamily: Dual specificity phosphatase-like
68 c4jmkA_
not modelled
14.0
21
PDB header: hydrolaseChain: A: PDB Molecule: dual specificity protein phosphatase 8;PDBTitle: structure of dusp8
69 c2aucC_
not modelled
14.0
7
PDB header: membrane proteinChain: C: PDB Molecule: myosin a tail interacting protein;PDBTitle: structure of the plasmodium mtip-myoa complex, a key component of the2 parasite invasion motor
70 d1hdgo1
not modelled
13.9
14
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain
71 d2b5id2
not modelled
13.6
33
Fold: Complement control module/SCR domainSuperfamily: Complement control module/SCR domainFamily: Complement control module/SCR domain
72 c2hcmA_
not modelled
13.6
30
PDB header: hydrolaseChain: A: PDB Molecule: dual specificity protein phosphatase;PDBTitle: crystal structure of mouse putative dual specificity phosphatase2 complexed with zinc tungstate, new york structural genomics3 consortium
73 c5xjvA_
not modelled
13.5
21
PDB header: hydrolaseChain: A: PDB Molecule: dual specificity protein phosphatase 13 isoform a;PDBTitle: two intermediate states of conformation switch in dual specificity2 phosphatase 13a
74 d1ggaa1
not modelled
13.4
26
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain
75 d1yq2a2
not modelled
13.4
17
Fold: Immunoglobulin-like beta-sandwichSuperfamily: beta-Galactosidase/glucuronidase domainFamily: beta-Galactosidase/glucuronidase domain
76 c2vfwB_
not modelled
13.1
44
PDB header: transferaseChain: B: PDB Molecule: short-chain z-isoprenyl diphosphate synthetase;PDBTitle: rv1086 native
77 c3ihtB_
not modelled
13.1
80
PDB header: transferaseChain: B: PDB Molecule: s-adenosyl-l-methionine methyl transferase;PDBTitle: crystal structure of s-adenosyl-l-methionine methyl transferase2 (yp_165822.1) from silicibacter pomeroyi dss-3 at 1.80 a resolution
78 d1d5ra2
not modelled
13.1
24
Fold: (Phosphotyrosine protein) phosphatases IISuperfamily: (Phosphotyrosine protein) phosphatases IIFamily: Dual specificity phosphatase-like
79 c1yz4A_
not modelled
13.0
25
PDB header: hydrolaseChain: A: PDB Molecule: dual specificity phosphatase-like 15 isoform a;PDBTitle: crystal structure of dusp15
80 d1i32a1
not modelled
13.0
22
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain
81 c5wolA_
not modelled
12.9
24
PDB header: oxidoreductaseChain: A: PDB Molecule: 4-hydroxy-tetrahydrodipicolinate reductase;PDBTitle: crystal structure of dihydrodipicolinate reductase dapb from coxiella2 burnetii
82 c4hd5A_
not modelled
12.9
0
PDB header: hydrolaseChain: A: PDB Molecule: polysaccharide deacetylase;PDBTitle: crystal structure of bc0361, a polysaccharide deacetylase from2 bacillus cereus
83 d1vpda1
not modelled
12.8
13
Fold: 6-phosphogluconate dehydrogenase C-terminal domain-likeSuperfamily: 6-phosphogluconate dehydrogenase C-terminal domain-likeFamily: Hydroxyisobutyrate and 6-phosphogluconate dehydrogenase domain
84 d3gpdg1
not modelled
12.6
18
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain
85 d1m3ga_
not modelled
12.5
26
Fold: (Phosphotyrosine protein) phosphatases IISuperfamily: (Phosphotyrosine protein) phosphatases IIFamily: Dual specificity phosphatase-like
86 c1yx7A_
not modelled
12.4
12
PDB header: metal binding proteinChain: A: PDB Molecule: calsensin;PDBTitle: nmr structure of calsensin, energy minimized average2 structure.
87 c6dzpg_
not modelled
12.0
57
PDB header: ribosomeChain: G: PDB Molecule: 50s ribosomal protein l6;PDBTitle: cryo-em structure of mycobacterium smegmatis c(minus) 50s ribosomal2 subunit
88 c5eesA_
not modelled
12.0
19
PDB header: oxidoreductaseChain: A: PDB Molecule: 4-hydroxy-tetrahydrodipicolinate reductase;PDBTitle: crystal strcuture of dapb in complex with nadp+ from corynebacterium2 glutamicum
89 c2vg2C_
not modelled
11.9
50
PDB header: transferaseChain: C: PDB Molecule: undecaprenyl pyrophosphate synthetase;PDBTitle: rv2361 with ipp
90 c1drwA_
not modelled
11.8
33
PDB header: oxidoreductaseChain: A: PDB Molecule: dihydrodipicolinate reductase;PDBTitle: escherichia coli dhpr/nhdh complex
91 c3h2zA_
not modelled
11.8
28
PDB header: oxidoreductaseChain: A: PDB Molecule: mannitol-1-phosphate 5-dehydrogenase;PDBTitle: the crystal structure of mannitol-1-phosphate dehydrogenase from2 shigella flexneri
92 d1vhra_
not modelled
11.4
26
Fold: (Phosphotyrosine protein) phosphatases IISuperfamily: (Phosphotyrosine protein) phosphatases IIFamily: Dual specificity phosphatase-like
93 d1k3ta1
not modelled
11.3
26
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain
94 d2cvza1
not modelled
11.2
9
Fold: 6-phosphogluconate dehydrogenase C-terminal domain-likeSuperfamily: 6-phosphogluconate dehydrogenase C-terminal domain-likeFamily: Hydroxyisobutyrate and 6-phosphogluconate dehydrogenase domain
95 c5xw4A_
not modelled
10.7
25
PDB header: cell cycleChain: A: PDB Molecule: tyrosine-protein phosphatase cdc14;PDBTitle: crystal structure of budding yeast cdc14p (wild type) in the apo state
96 c2j034_
not modelled
10.7
56
PDB header: ribosomeChain: 4: PDB Molecule: 50s ribosomal protein l31;PDBTitle: structure of the thermus thermophilus 70s ribosome2 complexed with mrna, trna and paromomycin (part 4 of 4).3 this file contains the 50s subunit from molecule ii.
97 d2j0141
not modelled
10.7
56
Fold: L28p-likeSuperfamily: L28p-likeFamily: Ribosomal protein L31p
98 c4q2uM_
not modelled
10.4
11
PDB header: toxin/toxin repressorChain: M: PDB Molecule: antitoxin dinj;PDBTitle: crystal structure of the e. coli dinj-yafq toxin-antitoxin complex
99 c5xk9F_
not modelled
10.4
25
PDB header: transferaseChain: F: PDB Molecule: undecaprenyl diphosphate synthase;PDBTitle: crystal structure of isosesquilavandulyl diphosphate synthase from2 streptomyces sp. strain cnh-189 in complex with gspp and dmapp