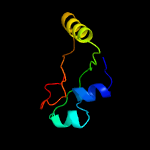
1 c2g38A_
99.8
35
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: pe family protein;PDBTitle: a pe/ppe protein complex from mycobacterium tuberculosis
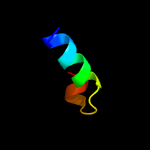
2 d2g38a1
99.8
35
Fold: Ferritin-likeSuperfamily: PE/PPE dimer-likeFamily: PE
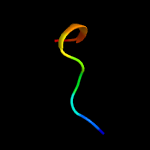
3 c5xfsA_
99.8
45
PDB header: protein transportChain: A: PDB Molecule: pe family protein pe8;PDBTitle: crystal structure of pe8-ppe15 in complex with espg5 from m.2 tuberculosis
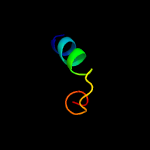
4 c2knpA_
41.5
56
PDB header: unknown functionChain: A: PDB Molecule: mcocc-1;PDBTitle: isolation and characterization of peptides from momordica2 cochinchinensis seeds.
5 c4uvkA_
21.0
19
PDB header: cell cycleChain: A: PDB Molecule: zyro0d15994p;PDBTitle: cohesin subunit scc3 from z. rouxii, 88-1035
6 c2kukA_
15.8
67
PDB header: antiviral proteinChain: A: PDB Molecule: leaf cyclotide 2;PDBTitle: solution structure of vhl-2
7 d1v29a_
15.8
58
Fold: Nitrile hydratase alpha chainSuperfamily: Nitrile hydratase alpha chainFamily: Nitrile hydratase alpha chain
8 c5sxpG_
15.5
78
PDB header: signaling protein/ligaseChain: G: PDB Molecule: e3 ubiquitin-protein ligase itchy homolog;PDBTitle: structural basis for the interaction between itch prr and beta-pix
9 d1w1oa2
14.9
24
Fold: FAD-binding/transporter-associated domain-likeSuperfamily: FAD-binding/transporter-associated domain-likeFamily: FAD-linked oxidases, N-terminal domain
10 c4pjwA_
14.9
29
PDB header: cell cycleChain: A: PDB Molecule: cohesin subunit sa-2;PDBTitle: crystal structure of human stromal antigen 2 (sa2) in complex with2 sister chromatid cohesion protein 1 (scc1), with bound mes
11 c1wlpA_
13.2
78
PDB header: oxidoreductase/signaling proteinChain: A: PDB Molecule: cytochrome b-245 light chain;PDBTitle: solution structure of the p22phox-p47phox complex
12 c6h8qA_
13.0
19
PDB header: cell cycleChain: A: PDB Molecule: cohesin subunit scc3;PDBTitle: structural basis for scc3-dependent cohesin recruitment to chromatin
13 c3gvmA_
11.2
22
PDB header: viral proteinChain: A: PDB Molecule: putative uncharacterized protein sag1039;PDBTitle: structure of the homodimeric wxg-100 family protein from streptococcus2 agalactiae
14 c6o58H_
10.8
33
PDB header: transport proteinChain: H: PDB Molecule: essential mcu regulator, mitochondrial;PDBTitle: human mcu-emre complex, dimer of channel
15 c6o58P_
10.8
33
PDB header: transport proteinChain: P: PDB Molecule: essential mcu regulator, mitochondrial;PDBTitle: human mcu-emre complex, dimer of channel
16 c5yixB_
10.6
38
PDB header: dna binding proteinChain: B: PDB Molecule: cell cycle regulatory protein gcra;PDBTitle: caulobacter crescentus gcra sigma-interacting domain (sid) in complex2 with domain 2 of sigma 70
17 d1f0la3
10.6
33
Fold: Toxins' membrane translocation domainsSuperfamily: Diphtheria toxin, middle domainFamily: Diphtheria toxin, middle domain
18 c2mn1A_
9.5
50
PDB header: unknown functionChain: A: PDB Molecule: kalata b1[w23ww];PDBTitle: solution structure of kalata b1[w23ww]
19 d1w0ba_
9.5
40
Fold: Spectrin repeat-likeSuperfamily: Alpha-hemoglobin stabilizing protein AHSPFamily: Alpha-hemoglobin stabilizing protein AHSP
20 d1mx3a2
9.4
31
Fold: Flavodoxin-likeSuperfamily: Formate/glycerate dehydrogenase catalytic domain-likeFamily: Formate/glycerate dehydrogenases, substrate-binding domain
21 d1eaic_
not modelled
9.1
50
Fold: Serine protease inhibitorsSuperfamily: Serine protease inhibitorsFamily: ATI-like
22 d1z8ua1
not modelled
9.0
42
Fold: Spectrin repeat-likeSuperfamily: Alpha-hemoglobin stabilizing protein AHSPFamily: Alpha-hemoglobin stabilizing protein AHSP
23 c5jhoA_
not modelled
8.3
13
PDB header: transport proteinChain: A: PDB Molecule: electroneutral sodium bicarbonate exchanger 1;PDBTitle: crystal structure of the regulatory domain of the sodium driven2 chloride bicarbonate exchanger.
24 c4lwsB_
not modelled
6.3
11
PDB header: unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: esxa : esxb (semet) hetero-dimer from thermomonospora curvata
25 c3bboG_
not modelled
5.6
40
PDB header: ribosomeChain: G: PDB Molecule: ribosomal protein l4;PDBTitle: homology model for the spinach chloroplast 50s subunit fitted to 9.4a2 cryo-em map of the 70s chlororibosome