| 1 |
|
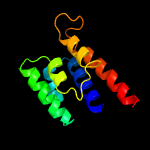
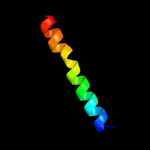
PDB 3c1q chain A
Region: 90 - 201
Aligned: 106
Modelled: 112
Confidence: 99.2%
Identity: 14%
PDB header:transport protein
Chain: A: PDB Molecule:general secretion pathway protein f;
PDBTitle: the three-dimensional structure of the cytoplasmic domains of epsf2 from the type 2 secretion system of vibrio cholerae
Phyre2
| 2 |
|
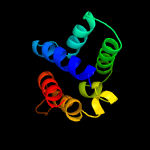
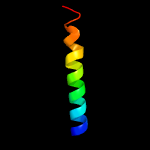
PDB 5nbg chain A
Region: 90 - 198
Aligned: 103
Modelled: 109
Confidence: 99.2%
Identity: 15%
PDB header:membrane protein
Chain: A: PDB Molecule:general secretion pathway protein f;
PDBTitle: structure of the cytoplasmic domain i of outf in the d. dadantii type2 ii secretion system
Phyre2
| 3 |
|
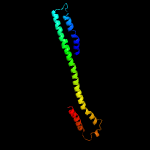
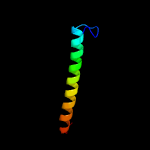
PDB 2whn chain A
Region: 90 - 194
Aligned: 98
Modelled: 105
Confidence: 98.3%
Identity: 11%
PDB header:protein transport
Chain: A: PDB Molecule:pilus assembly protein pilc;
PDBTitle: n-terminal domain from the pilc type iv pilus biogenesis protein
Phyre2
| 4 |
|
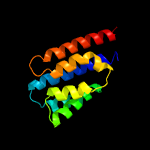
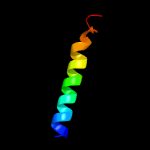
PDB 4hhx chain A
Region: 90 - 187
Aligned: 89
Modelled: 98
Confidence: 97.3%
Identity: 13%
PDB header:membrane protein
Chain: A: PDB Molecule:toxin coregulated pilus biosynthesis protein e;
PDBTitle: structure of cytoplasmic domain of tcpe from vibrio cholerae
Phyre2
| 5 |
|
PDB 5n9y chain B
Region: 125 - 259
Aligned: 133
Modelled: 135
Confidence: 89.1%
Identity: 12%
PDB header:membrane protein
Chain: B: PDB Molecule:zinc transport protein zntb;
PDBTitle: the full-length structure of zntb
Phyre2
| 6 |
|
PDB 6bs9 chain A
Region: 82 - 196
Aligned: 113
Modelled: 115
Confidence: 82.2%
Identity: 19%
PDB header:transport protein
Chain: A: PDB Molecule:stage iii sporulation protein ab;
PDBTitle: stage iii sporulation protein ab (spoiiiab)
Phyre2
| 7 |
|
PDB 4i0x chain A
Region: 89 - 155
Aligned: 67
Modelled: 67
Confidence: 35.0%
Identity: 22%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:esat-6-like protein mab_3112;
PDBTitle: crystal structure of the mycobacterum abscessus esxef (mab_3112-2 mab_3113) complex
Phyre2
| 8 |
|
PDB 4ev6 chain E
Region: 84 - 259
Aligned: 165
Modelled: 171
Confidence: 32.5%
Identity: 13%
PDB header:metal transport
Chain: E: PDB Molecule:magnesium transport protein cora;
PDBTitle: the complete structure of cora magnesium transporter from2 methanocaldococcus jannaschii
Phyre2
| 9 |
|
PDB 5eze chain G
Region: 174 - 201
Aligned: 28
Modelled: 28
Confidence: 27.9%
Identity: 18%
PDB header:de novo protein
Chain: G: PDB Molecule:cc-hept-bmecys-his-glu;
PDBTitle: a de novo designed heptameric coiled coil cc-hept-i18betamecys-l22h-2 i25e
Phyre2
| 10 |
|
PDB 5eze chain F
Region: 174 - 199
Aligned: 26
Modelled: 26
Confidence: 23.0%
Identity: 19%
PDB header:de novo protein
Chain: F: PDB Molecule:cc-hept-bmecys-his-glu;
PDBTitle: a de novo designed heptameric coiled coil cc-hept-i18betamecys-l22h-2 i25e
Phyre2
| 11 |
|
PDB 5f2y chain A
Region: 174 - 199
Aligned: 26
Modelled: 26
Confidence: 23.0%
Identity: 19%
PDB header:de novo protein
Chain: A: PDB Molecule:cc-hept-hcys-h-e;
PDBTitle: a de novo designed heptameric coiled coil cc-hept-homocys-h-e
Phyre2
| 12 |
|
PDB 5eze chain A
Region: 174 - 199
Aligned: 26
Modelled: 26
Confidence: 23.0%
Identity: 19%
PDB header:de novo protein
Chain: A: PDB Molecule:cc-hept-bmecys-his-glu;
PDBTitle: a de novo designed heptameric coiled coil cc-hept-i18betamecys-l22h-2 i25e
Phyre2
| 13 |
|
PDB 5eze chain E
Region: 174 - 199
Aligned: 26
Modelled: 26
Confidence: 22.9%
Identity: 19%
PDB header:de novo protein
Chain: E: PDB Molecule:cc-hept-bmecys-his-glu;
PDBTitle: a de novo designed heptameric coiled coil cc-hept-i18betamecys-l22h-2 i25e
Phyre2
| 14 |
|
PDB 5f2y chain E
Region: 174 - 199
Aligned: 26
Modelled: 26
Confidence: 22.9%
Identity: 19%
PDB header:de novo protein
Chain: E: PDB Molecule:cc-hept-hcys-h-e;
PDBTitle: a de novo designed heptameric coiled coil cc-hept-homocys-h-e
Phyre2
| 15 |
|
PDB 5eze chain C
Region: 174 - 199
Aligned: 26
Modelled: 26
Confidence: 22.9%
Identity: 19%
PDB header:de novo protein
Chain: C: PDB Molecule:cc-hept-bmecys-his-glu;
PDBTitle: a de novo designed heptameric coiled coil cc-hept-i18betamecys-l22h-2 i25e
Phyre2
| 16 |
|
PDB 6a6x chain C
Region: 169 - 209
Aligned: 41
Modelled: 41
Confidence: 22.7%
Identity: 17%
PDB header:toxin
Chain: C: PDB Molecule:antitoxin maze7;
PDBTitle: the crystal structure of the mtb maze-mazf-mt9 complex
Phyre2
| 17 |
|
PDB 5f2y chain G
Region: 174 - 199
Aligned: 26
Modelled: 26
Confidence: 22.5%
Identity: 19%
PDB header:de novo protein
Chain: G: PDB Molecule:cc-hept-hcys-h-e;
PDBTitle: a de novo designed heptameric coiled coil cc-hept-homocys-h-e
Phyre2
| 18 |
|
PDB 5f2y chain F
Region: 174 - 196
Aligned: 23
Modelled: 23
Confidence: 10.9%
Identity: 17%
PDB header:de novo protein
Chain: F: PDB Molecule:cc-hept-hcys-h-e;
PDBTitle: a de novo designed heptameric coiled coil cc-hept-homocys-h-e
Phyre2
| 19 |
|
PDB 5f2y chain B
Region: 174 - 196
Aligned: 23
Modelled: 23
Confidence: 10.0%
Identity: 17%
PDB header:de novo protein
Chain: B: PDB Molecule:cc-hept-hcys-h-e;
PDBTitle: a de novo designed heptameric coiled coil cc-hept-homocys-h-e
Phyre2
| 20 |
|
PDB 3l8j chain A
Region: 93 - 205
Aligned: 108
Modelled: 113
Confidence: 8.1%
Identity: 17%
PDB header:protein binding
Chain: A: PDB Molecule:programmed cell death protein 10;
PDBTitle: crystal structure of ccm3, a cerebral cavernous malformation protein2 critical for vascular integrity
Phyre2
| 21 |
|