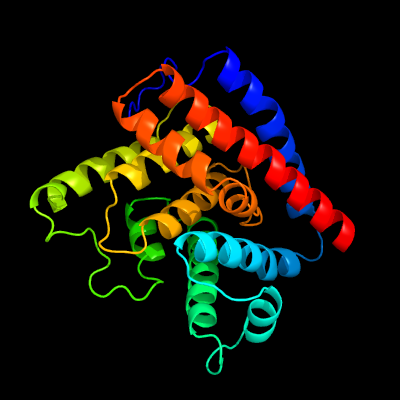
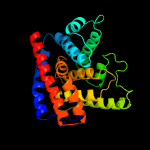
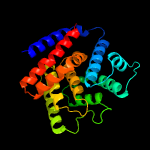
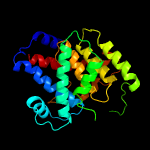
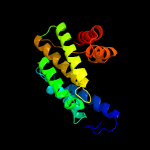
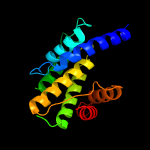
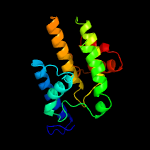
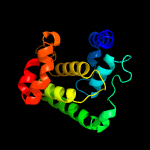
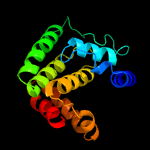
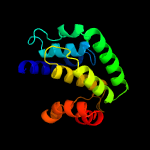
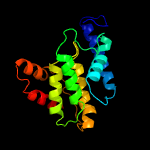
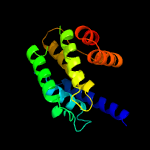
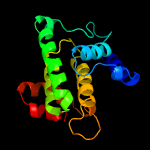
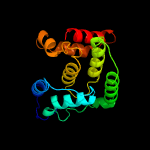
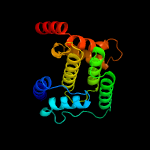
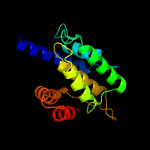
1 c3eqxB_
100.0
16
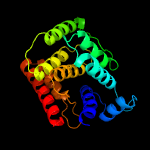
PDB header: dna binding proteinChain: B: PDB Molecule: fic domain containing transcriptional regulator;PDBTitle: crystal structure of a fic family protein (so_4266) from shewanella2 oneidensis at 1.6 a resolution
2 c4rglA_
100.0
20
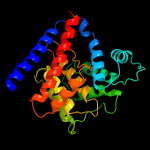
PDB header: dna binding proteinChain: A: PDB Molecule: filamentation induced by camp protein fic;PDBTitle: crystal structure of a fic family protein (dde_2494) from2 desulfovibrio desulfuricans g20 at 2.70 a resolution
3 c3cucB_
100.0
13
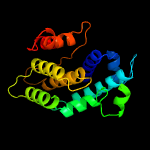
PDB header: signaling proteinChain: B: PDB Molecule: protein of unknown function with a fic domain;PDBTitle: crystal structure of a fic domain containing signaling protein2 (bt_2513) from bacteroides thetaiotaomicron vpi-5482 at 2.71 a3 resolution
4 c4u0zH_
100.0
15
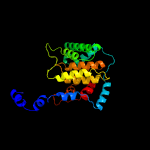
PDB header: transferaseChain: H: PDB Molecule: adenosine monophosphate-protein transferase ficd;PDBTitle: eukaryotic fic domain containing protein with bound apcpp
5 c5jj6A_
100.0
16
PDB header: transferaseChain: A: PDB Molecule: adenosine monophosphate-protein transferase ficd homolog;PDBTitle: fic-1 (aa134 - 508) from c. elegans
6 c4x2eA_
100.0
12
PDB header: transferaseChain: A: PDB Molecule: fic family protein putative filamentation induced by campPDBTitle: clostridium difficile wild type fic protein
7 c3n3vA_
99.9
10
PDB header: transferaseChain: A: PDB Molecule: adenosine monophosphate-protein transferase ibpa;PDBTitle: crystal structure of ibpafic2-h3717a in complex with adenylylated2 cdc42
8 c5jffC_
99.9
15
PDB header: transferaseChain: C: PDB Molecule: probable adenosine monophosphate-protein transferase fic;PDBTitle: e. coli ecfict mutant g55r in complex with ecfica
9 c4lu4A_
99.9
13
PDB header: transferaseChain: A: PDB Molecule: putative cell filamentation protein;PDBTitle: crystal structure of the n-terminal fic domain of a putative cell2 filamentation protein (virb-translocated bep effector protein) from3 bartonella quintana
10 c3shgA_
99.8
13
PDB header: transferase/protein bindingChain: A: PDB Molecule: vbht;PDBTitle: vbht fic protein from bartonella schoenbuchensis in complex with vbha2 antitoxin
11 c2f6sA_
99.8
12
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: cell filamentation protein, putative;PDBTitle: structure of cell filamentation protein (fic) from helicobacter pylori
12 c5nwfB_
99.8
12
PDB header: toxinChain: B: PDB Molecule: fic family protein;PDBTitle: enterococcus faecalis fic protein (h111a).
13 d2f6sa1
99.8
12
Fold: Fic-likeSuperfamily: Fic-likeFamily: Fic-like
14 c4xi8B_
99.8
11
PDB header: protein bindingChain: B: PDB Molecule: bartonella effector protein (bep) substrate of virb t4ss;PDBTitle: crystal structure of the fic domain of bep5 protein (virb-translocated2 bartonella effector protein) from bartonella clarridgeiae
15 c4py3B_
99.8
11
PDB header: protein bindingChain: B: PDB Molecule: bartonella effector protein (bep) substrate of virb t4ss;PDBTitle: crystal structure of the n-terminal fic domain of bep8 protein (virb-2 translocated bartonella effector protein) from bartonella sp. 1-1c
16 d2g03a1
99.8
13
Fold: Fic-likeSuperfamily: Fic-likeFamily: Fic-like
17 c4m16A_
99.7
9
PDB header: cell adhesionChain: A: PDB Molecule: bartonella effector protein (bep) substrate of virb t4ss;PDBTitle: crystal structure of the n-terminal fic domain of bartonella effector2 protein (bep); substrate of virb t4ss (virb-translocated bep effector3 protein) from bartonella sp. ar 15-3
18 c4npsA_
99.7
12
PDB header: cell adhesionChain: A: PDB Molecule: bartonella effector protein (bep) substrate of virb t4ss;PDBTitle: crystal structure of bep1 protein (virb-translocated bartonella2 effector protein) from bartonella clarridgeiae
19 c2vzaD_
99.4
14
PDB header: cell adhesionChain: D: PDB Molecule: cell filamentation protein;PDBTitle: type iv secretion system effector protein bepa
20 c3dd7A_
98.8
17
PDB header: ribosome inhibitorChain: A: PDB Molecule: death on curing protein;PDBTitle: structure of doch66y in complex with the c-terminal domain of phd
21 c3letB_
not modelled
98.3
10
PDB header: transferaseChain: B: PDB Molecule: adenosine monophosphate-protein transferase vops;PDBTitle: crystal structure of fic domain containing ampylator, vops
22 d1v54h_
not modelled
50.0
11
Fold: Cytochrome c oxidase subunit hSuperfamily: Cytochrome c oxidase subunit hFamily: Cytochrome c oxidase subunit h
23 c6h1nA_
not modelled
40.9
16
PDB header: apoptosisChain: A: PDB Molecule: bcl2-like 10 (apoptosis facilitator);PDBTitle: crystal structure of a zebra-fish pro-survival protein nrz-apo
24 d1yl7a1
not modelled
34.2
37
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain
25 d1u8fo1
not modelled
28.5
21
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain
26 d1j0xo1
not modelled
28.0
21
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain
27 d1diha1
not modelled
25.6
21
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain
28 d1ggaa1
not modelled
24.5
18
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain
29 d3gpdg1
not modelled
24.1
21
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain
30 d1s2xa_
not modelled
23.6
27
Fold: STAT-likeSuperfamily: Cag-ZFamily: Cag-Z
31 c1s2xA_
not modelled
23.6
27
PDB header: unknown functionChain: A: PDB Molecule: cag-z;PDBTitle: crystal structure of cag-z from helicobacter pylori
32 d1k3ta1
not modelled
23.0
18
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain
33 c3ijpA_
not modelled
22.7
16
PDB header: oxidoreductaseChain: A: PDB Molecule: dihydrodipicolinate reductase;PDBTitle: crystal structure of dihydrodipicolinate reductase from bartonella2 henselae at 2.0a resolution
34 d2b4ro1
not modelled
22.3
21
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain
35 d1s0pa_
not modelled
20.4
12
Fold: N-terminal domain of adenylylcyclase associated protein, CAPSuperfamily: N-terminal domain of adenylylcyclase associated protein, CAPFamily: N-terminal domain of adenylylcyclase associated protein, CAP
36 d1hdgo1
not modelled
20.3
25
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain
37 d1gado1
not modelled
19.7
29
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain
38 c1yl7F_
not modelled
19.3
37
PDB header: oxidoreductaseChain: F: PDB Molecule: dihydrodipicolinate reductase;PDBTitle: the crystal structure of mycobacterium tuberculosis2 dihydrodipicolinate reductase (rv2773c) in complex with nadh (crystal3 form c)
39 d1obfo1
not modelled
19.0
21
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain
40 c4aezC_
not modelled
18.4
10
PDB header: cell cycleChain: C: PDB Molecule: mitotic spindle checkpoint component mad3;PDBTitle: crystal structure of mitotic checkpoint complex
41 c3idwA_
not modelled
18.0
13
PDB header: endocytosisChain: A: PDB Molecule: actin cytoskeleton-regulatory complex protein sla1;PDBTitle: crystal structure of sla1 homology domain 2
42 c5lcwS_
not modelled
17.1
12
PDB header: cell cycleChain: S: PDB Molecule: mitotic checkpoint serine/threonine-protein kinase bub1PDBTitle: cryo-em structure of the anaphase-promoting complex/cyclosome, in2 complex with the mitotic checkpoint complex (apc/c-mcc) at 4.23 angstrom resolution
43 c1d5rA_
not modelled
16.9
9
PDB header: hydrolaseChain: A: PDB Molecule: phosphoinositide phosphatase pten;PDBTitle: crystal structure of the pten tumor suppressor
44 d1vc2a1
not modelled
15.8
26
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain
45 c2p58C_
not modelled
15.7
22
PDB header: transport protein/chaperoneChain: C: PDB Molecule: putative type iii secretion protein yscg;PDBTitle: structure of the yersinia pestis type iii secretion system needle2 protein yscf in complex with its chaperones ysce/yscg
46 d1rm4a1
not modelled
15.5
17
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain
47 c5kt0A_
not modelled
15.2
32
PDB header: oxidoreductaseChain: A: PDB Molecule: 4-hydroxy-tetrahydrodipicolinate reductase;PDBTitle: dihydrodipicolinate reductase from the industrial and evolutionarily2 important cyanobacteria anabaena variabilis.
48 c5eesA_
not modelled
14.8
26
PDB header: oxidoreductaseChain: A: PDB Molecule: 4-hydroxy-tetrahydrodipicolinate reductase;PDBTitle: crystal strcuture of dapb in complex with nadp+ from corynebacterium2 glutamicum
49 c5wolA_
not modelled
14.8
26
PDB header: oxidoreductaseChain: A: PDB Molecule: 4-hydroxy-tetrahydrodipicolinate reductase;PDBTitle: crystal structure of dihydrodipicolinate reductase dapb from coxiella2 burnetii
50 d1vm6a3
not modelled
14.6
8
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain
51 d3cmco1
not modelled
14.6
21
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain
52 c6dzpg_
not modelled
13.9
29
PDB header: ribosomeChain: G: PDB Molecule: 50s ribosomal protein l6;PDBTitle: cryo-em structure of mycobacterium smegmatis c(minus) 50s ribosomal2 subunit
53 d1dssg1
not modelled
13.6
25
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain
54 d3etja2
not modelled
12.7
22
Fold: PreATP-grasp domainSuperfamily: PreATP-grasp domainFamily: BC N-terminal domain-like
55 d2pkqo1
not modelled
12.6
21
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain
56 c5ugjC_
not modelled
12.5
18
PDB header: oxidoreductaseChain: C: PDB Molecule: 4-hydroxy-tetrahydrodipicolinate reductase;PDBTitle: crystal structure of htpa reductase from neisseria meningitidis
57 c1i32D_
not modelled
12.4
14
PDB header: oxidoreductaseChain: D: PDB Molecule: glyceraldehyde 3-phosphate dehydrogenase;PDBTitle: leishmania mexicana glyceraldehyde-3-phosphate2 dehydrogenase in complex with inhibitors
58 d1i32a1
not modelled
12.2
14
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain
59 d1uzxa_
not modelled
12.1
10
Fold: UBC-likeSuperfamily: UBC-likeFamily: UEV domain
60 c2j034_
not modelled
12.0
22
PDB header: ribosomeChain: 4: PDB Molecule: 50s ribosomal protein l31;PDBTitle: structure of the thermus thermophilus 70s ribosome2 complexed with mrna, trna and paromomycin (part 4 of 4).3 this file contains the 50s subunit from molecule ii.
61 d2j0141
not modelled
12.0
22
Fold: L28p-likeSuperfamily: L28p-likeFamily: Ribosomal protein L31p
62 c3hq4R_
not modelled
11.5
26
PDB header: oxidoreductaseChain: R: PDB Molecule: glyceraldehyde-3-phosphate dehydrogenase 1;PDBTitle: crystal structure of c151s mutant of glyceraldehyde-3-phosphate2 dehydrogenase 1 (gapdh1) complexed with nad from staphylococcus3 aureus mrsa252 at 2.2 angstrom resolution
63 c3bhwA_
not modelled
11.3
9
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of an uncharacterized protein from magnetospirillum2 magneticum
64 d1vs6z1
not modelled
11.1
20
Fold: L28p-likeSuperfamily: L28p-likeFamily: Ribosomal protein L31p
65 c5ld5C_
not modelled
11.1
21
PDB header: oxidoreductaseChain: C: PDB Molecule: glyceraldehyde-3-phosphate dehydrogenase;PDBTitle: crystal structure of a bacterial dehydrogenase at 2.19 angstroms2 resolution
66 d1bf4a_
not modelled
10.4
20
Fold: SH3-like barrelSuperfamily: Chromo domain-likeFamily: "Histone-like" proteins from archaea
67 c2ksdA_
not modelled
10.3
11
PDB header: transferaseChain: A: PDB Molecule: aerobic respiration control sensor protein arcb;PDBTitle: backbone structure of the membrane domain of e. coli histidine kinase2 receptor arcb, center for structures of membrane proteins (csmp)3 target 4310c
68 c6hu9j_
not modelled
10.3
16
PDB header: oxidoreductase/electron transportChain: J: PDB Molecule: cytochrome b-c1 complex subunit 10;PDBTitle: iii2-iv2 mitochondrial respiratory supercomplex from s. cerevisiae
69 d2g82a1
not modelled
9.9
25
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain
70 c1hdgO_
not modelled
9.7
25
PDB header: oxidoreductase (aldehy(d)-nad(a))Chain: O: PDB Molecule: holo-d-glyceraldehyde-3-phosphate dehydrogenase;PDBTitle: the crystal structure of holo-glyceraldehyde-3-phosphate dehydrogenase2 from the hyperthermophilic bacterium thermotoga maritima at 2.53 angstroms resolution
71 c2yyyB_
not modelled
9.7
17
PDB header: oxidoreductaseChain: B: PDB Molecule: glyceraldehyde-3-phosphate dehydrogenase;PDBTitle: crystal structure of glyceraldehyde-3-phosphate2 dehydrogenase
72 c2ep7B_
not modelled
9.5
21
PDB header: oxidoreductaseChain: B: PDB Molecule: glyceraldehyde-3-phosphate dehydrogenase;PDBTitle: structural study of project id aq_1065 from aquifex aeolicus vf5
73 c6g21B_
not modelled
9.4
7
PDB header: hydrolaseChain: B: PDB Molecule: probable feruloyl esterase b-2;PDBTitle: crystal structure of an esterase from aspergillus oryzae
74 c6fqbE_
not modelled
9.4
12
PDB header: ligaseChain: E: PDB Molecule: cobyric acid synthase;PDBTitle: murt/gatd peptidoglycan amidotransferase complex from streptococcus2 pneumoniae r6
75 c1drwA_
not modelled
9.4
21
PDB header: oxidoreductaseChain: A: PDB Molecule: dihydrodipicolinate reductase;PDBTitle: escherichia coli dhpr/nhdh complex
76 d2j01d1
not modelled
9.0
30
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: C-terminal domain of ribosomal protein L2
77 c5ur0B_
not modelled
9.0
29
PDB header: oxidoreductaseChain: B: PDB Molecule: glyceraldehyde-3-phosphate dehydrogenase;PDBTitle: crystallographic structure of glyceraldehyde-3-phosphate dehydrogenase2 from naegleria gruberi
78 c3pk1A_
not modelled
9.0
13
PDB header: apoptosis/apoptosis regulatorChain: A: PDB Molecule: induced myeloid leukemia cell differentiation protein mcl-PDBTitle: crystal structure of mcl-1 in complex with the baxbh3 domain
79 c4p40A_
not modelled
8.8
9
PDB header: transport proteinChain: A: PDB Molecule: copn;PDBTitle: chlamydia pneumoniae copn
80 c3wmtA_
not modelled
8.8
7
PDB header: hydrolaseChain: A: PDB Molecule: probable feruloyl esterase b-1;PDBTitle: crystal structure of feruloyl esterase b from aspergillus oryzae
81 c3nfqA_
not modelled
8.7
15
PDB header: transcriptionChain: A: PDB Molecule: transcription factor iws1;PDBTitle: crystal structure of the conserved central domain of yeast spn1/iws1
82 d1adta1
not modelled
8.7
5
Fold: Domain of early E2A DNA-binding protein, ADDBPSuperfamily: Domain of early E2A DNA-binding protein, ADDBPFamily: Domain of early E2A DNA-binding protein, ADDBP
83 c4qx6A_
not modelled
8.6
21
PDB header: oxidoreductaseChain: A: PDB Molecule: glyceraldehyde 3-phosphate dehydrogenase;PDBTitle: crystal structure of glyceraldehyde-3-phosphate dehydrogenase from2 streptococcus agalactiae nem316 at 2.46 angstrom resolution
84 c3lhlA_
not modelled
8.5
20
PDB header: hydrolaseChain: A: PDB Molecule: putative agmatinase;PDBTitle: crystal structure of a putative agmatinase from clostridium difficile
85 c3cieC_
not modelled
8.4
29
PDB header: oxidoreductaseChain: C: PDB Molecule: glyceraldehyde-3-phosphate dehydrogenase;PDBTitle: crystal structure of glyceraldehyde 3-phosphate2 dehydrogenase from cryptosporidium parvum
86 d1v54g_
not modelled
8.4
17
Fold: Single transmembrane helixSuperfamily: Mitochondrial cytochrome c oxidase subunit VIaFamily: Mitochondrial cytochrome c oxidase subunit VIa
87 c3wg7T_
not modelled
8.4
17
PDB header: oxidoreductaseChain: T: PDB Molecule: cytochrome c oxidase subunit 6a2, mitochondrial;PDBTitle: a 1.9 angstrom radiation damage free x-ray structure of large (420kda)2 protein by femtosecond crystallography
88 c5z62G_
not modelled
8.3
33
PDB header: electron transportChain: G: PDB Molecule: cytochrome c oxidase subunit 6a1, mitochondrial;PDBTitle: structure of human cytochrome c oxidase
89 c3zd0A_
not modelled
8.3
18
PDB header: transport proteinChain: A: PDB Molecule: p7 protein;PDBTitle: the solution structure of monomeric hepatitis c virus p72 yields potent inhibitors of virion release
90 c2gd1P_
not modelled
8.1
29
PDB header: oxidoreductase(aldehyde(d)-nad(a))Chain: P: PDB Molecule: apo-d-glyceraldehyde-3-phosphate dehydrogenase;PDBTitle: coenzyme-induced conformational changes in glyceraldehyde-3-2 phosphate dehydrogenase from bacillus stearothermophillus
91 d1sw8a_
not modelled
8.0
20
Fold: EF Hand-likeSuperfamily: EF-handFamily: Calmodulin-like
92 c5jyfB_
not modelled
8.0
21
PDB header: oxidoreductaseChain: B: PDB Molecule: glyceraldehyde-3-phosphate dehydrogenase;PDBTitle: structures of streptococcus agalactiae gbs gapdh in different2 enzymatic states
93 c3b20R_
not modelled
8.0
20
PDB header: oxidoreductaseChain: R: PDB Molecule: glyceraldehyde 3-phosphate dehydrogenase (nadp+);PDBTitle: crystal structure of glyceraldehyde-3-phosphate dehydrogenase2 complexed with nadfrom synechococcus elongatus"
94 c4l8nA_
not modelled
7.9
38
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: pdz domain protein;PDBTitle: crystal structure of a pdz domain protein (bdi_1242) from2 parabacteroides distasonis atcc 8503 at 2.50 a resolution
95 c3h9eO_
not modelled
7.9
28
PDB header: oxidoreductaseChain: O: PDB Molecule: glyceraldehyde-3-phosphate dehydrogenase, testis-specific;PDBTitle: crystal structure of human sperm-specific glyceraldehyde-3-phosphate2 dehydrogenase (gapds) complex with nad and phosphate
96 c2vfwB_
not modelled
7.9
22
PDB header: transferaseChain: B: PDB Molecule: short-chain z-isoprenyl diphosphate synthetase;PDBTitle: rv1086 native
97 c2b664_
not modelled
7.7
29
PDB header: ribosomeChain: 4: PDB Molecule: 50s ribosomal protein l31;PDBTitle: 50s ribosomal subunit from a crystal structure of release factor rf1,2 trnas and mrna bound to the ribosome. this file contains the 50s3 subunit from a crystal structure of release factor rf1, trnas and4 mrna bound to the ribosome and is described in remark 400
98 c2b4rQ_
not modelled
7.7
21
PDB header: oxidoreductaseChain: Q: PDB Molecule: glyceraldehyde-3-phosphate dehydrogenase;PDBTitle: crystal structure of glyceraldehyde-3-phosphate dehydrogenase from2 plasmodium falciparum at 2.25 angstrom resolution reveals intriguing3 extra electron density in the active site
99 c4r7rA_
not modelled
7.6
45
PDB header: lipid binding proteinChain: A: PDB Molecule: putative lipoprotein;PDBTitle: crystal structure of putative lipoprotein from clostridium perfringens