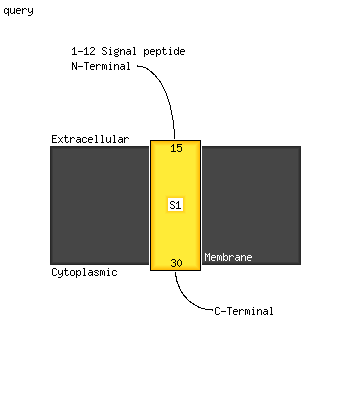
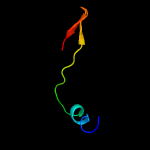
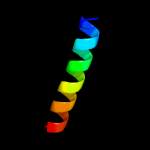
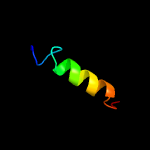
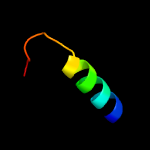
1 d1rfea_
40.5
18
Fold: Split barrel-likeSuperfamily: FMN-binding split barrelFamily: PNP-oxidase like
2 c6gqaD_
39.8
36
PDB header: cell cycleChain: D: PDB Molecule: cell cycle protein gpsb;PDBTitle: cell division regulator s. pneumoniae gpsb
3 c2wukD_
35.3
45
PDB header: cell cycleChain: D: PDB Molecule: septum site-determining protein diviva;PDBTitle: diviva n-terminal domain, f17a mutant
4 c4ug3C_
32.9
36
PDB header: cell cycleChain: C: PDB Molecule: cell cycle protein gpsb;PDBTitle: b. subtilis gpsb n-terminal domain
5 c4ug1A_
29.9
45
PDB header: cell cycleChain: A: PDB Molecule: cell cycle protein gpsb;PDBTitle: gpsb n-terminal domain
6 c3h36A_
29.6
53
PDB header: transferaseChain: A: PDB Molecule: polyribonucleotide nucleotidyltransferase;PDBTitle: structure of an uncharacterized domain in polyribonucleotide2 nucleotidyltransferase from streptococcus mutans ua159
7 c4fe1M_
15.5
31
PDB header: photosynthesisChain: M: PDB Molecule: photosystem i reaction center subunit xii;PDBTitle: improving the accuracy of macromolecular structure refinement at 7 a2 resolution
8 d1jb0m_
15.5
31
Fold: Single transmembrane helixSuperfamily: Subunit XII of photosystem I reaction centre, PsaMFamily: Subunit XII of photosystem I reaction centre, PsaM
9 c1jb0M_
15.5
31
PDB header: photosynthesisChain: M: PDB Molecule: photosystem 1 reaction centre subunit xii;PDBTitle: crystal structure of photosystem i: a photosynthetic reaction center2 and core antenna system from cyanobacteria
10 c5da9D_
15.3
24
PDB header: hydrolaseChain: D: PDB Molecule: putative double-strand break protein;PDBTitle: atp-gamma-s bound rad50 from chaetomium thermophilum in complex with2 the rad50-binding domain of mre11
11 c6g04A_
13.5
17
PDB header: transport proteinChain: A: PDB Molecule: pre-rrna-processing protein tsr2;PDBTitle: nmr solution structure of yeast tsr2(1-152) in complex with s26a(100-2 119)
12 d2py8a1
12.4
20
Fold: RbcX-likeSuperfamily: RbcX-likeFamily: RbcX-like
13 c5xj0Y_
12.2
24
PDB header: transferase/transcriptionChain: Y: PDB Molecule: gp76;PDBTitle: t. thermophilus rna polymerase holoenzyme bound with gp39 and gp76
14 d1w4ha1
12.1
38
Fold: Peripheral subunit-binding domain of 2-oxo acid dehydrogenase complexSuperfamily: Peripheral subunit-binding domain of 2-oxo acid dehydrogenase complexFamily: Peripheral subunit-binding domain of 2-oxo acid dehydrogenase complex
15 c3tgvD_
11.4
19
PDB header: heme binding proteinChain: D: PDB Molecule: heme-binding protein hutz;PDBTitle: crystal structure of hutz,the heme storsge protein from vibrio2 cholerae
16 c3f7eB_
11.1
19
PDB header: unknown functionChain: B: PDB Molecule: pyridoxamine 5'-phosphate oxidase-related, fmn-PDBTitle: msmeg_3380 f420 reductase
17 d1k4ta1
10.3
43
Fold: Long alpha-hairpinSuperfamily: Eukaryotic DNA topoisomerase I, dispensable insert domainFamily: Eukaryotic DNA topoisomerase I, dispensable insert domain
18 c2ksdA_
10.3
24
PDB header: transferaseChain: A: PDB Molecule: aerobic respiration control sensor protein arcb;PDBTitle: backbone structure of the membrane domain of e. coli histidine kinase2 receptor arcb, center for structures of membrane proteins (csmp)3 target 4310c
19 c1kmiZ_
9.9
10
PDB header: signaling proteinChain: Z: PDB Molecule: chemotaxis protein chez;PDBTitle: crystal structure of an e.coli chemotaxis protein, chez
20 d1j5ya2
9.6
19
Fold: HPr-likeSuperfamily: Putative transcriptional regulator TM1602, C-terminal domainFamily: Putative transcriptional regulator TM1602, C-terminal domain
21 c1pk1A_
not modelled
8.9
14
PDB header: transcription repressionChain: A: PDB Molecule: polyhomeotic-proximal chromatin protein;PDBTitle: hetero sam domain structure of ph and scm.
22 d2f66c1
not modelled
8.5
27
Fold: Long alpha-hairpinSuperfamily: Endosomal sorting complex assembly domainFamily: VPS37 C-terminal domain-like
23 c2hhzA_
not modelled
8.4
31
PDB header: oxidoreductaseChain: A: PDB Molecule: pyridoxamine 5'-phosphate oxidase-related;PDBTitle: crystal structure of a pyridoxamine 5'-phosphate oxidase-related2 protein (ssuidraft_2804) from streptococcus suis 89/1591 at 2.00 a3 resolution
24 d1w9aa_
not modelled
8.0
11
Fold: Split barrel-likeSuperfamily: FMN-binding split barrelFamily: PNP-oxidase like
25 c5j8yC_
not modelled
8.0
25
PDB header: nuclear proteinChain: C: PDB Molecule: polycomb protein sfmbt;PDBTitle: crystal structure of the scm-sam and sfmbt-sam heterodimer
26 c1pk1B_
not modelled
7.7
18
PDB header: transcription repressionChain: B: PDB Molecule: sex comb on midleg cg9495-pa;PDBTitle: hetero sam domain structure of ph and scm.
27 d1ldda_
not modelled
7.5
14
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: SCF ubiquitin ligase complex WHB domain
28 c4pzoE_
not modelled
7.5
14
PDB header: dna binding proteinChain: E: PDB Molecule: polyhomeotic-like protein 3;PDBTitle: crystal structure of phc3 sam l967r
29 c3bq7A_
not modelled
7.1
18
PDB header: transferaseChain: A: PDB Molecule: diacylglycerol kinase delta;PDBTitle: sam domain of diacylglycerol kinase delta1 (e35g)
30 d1pk1c1
not modelled
7.1
14
Fold: SAM domain-likeSuperfamily: SAM/Pointed domainFamily: SAM (sterile alpha motif) domain
31 d1pk3a1
not modelled
7.1
18
Fold: SAM domain-likeSuperfamily: SAM/Pointed domainFamily: SAM (sterile alpha motif) domain
32 d1z0jb1
not modelled
7.0
21
Fold: Long alpha-hairpinSuperfamily: Rabenosyn-5 Rab-binding domain-likeFamily: Rabenosyn-5 Rab-binding domain-like
33 c2dkzA_
not modelled
6.9
14
PDB header: signaling proteinChain: A: PDB Molecule: hypothetical protein loc64762;PDBTitle: solution structure of the sam_pnt-domain of the2 hypothetical protein loc64762
34 d1kw4a_
not modelled
6.9
14
Fold: SAM domain-likeSuperfamily: SAM/Pointed domainFamily: SAM (sterile alpha motif) domain
35 c3kfuJ_
not modelled
6.6
21
PDB header: ligase/rnaChain: J: PDB Molecule: glutamyl-trna(gln) amidotransferase subunit c;PDBTitle: crystal structure of the transamidosome
36 c5lkdB_
not modelled
6.6
32
PDB header: transferaseChain: B: PDB Molecule: glutathione s-transferase omega-like 2;PDBTitle: crystal structure of the xi glutathione transferase ecm4 from2 saccharomyces cerevisiae in complex with glutathione
37 c4qoyE_
not modelled
6.4
23
PDB header: oxidoreductaseChain: E: PDB Molecule: pyruvate dehydrogenase (dihydrolipoyltransacetylasePDBTitle: novel binding motif and new flexibility revealed by structural2 analysis of a pyruvate dehydrogenase-dihydrolipoyl acetyltransferase3 sub-complex from the escherichia coli pyruvate dehydrogenase multi-4 enzyme complex
38 d2af7a1
not modelled
6.3
21
Fold: AhpD-likeSuperfamily: AhpD-likeFamily: CMD-like
39 c3iz5n_
not modelled
5.9
25
PDB header: ribosomeChain: N: PDB Molecule: 60s ribosomal protein l14 (l14e);PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
40 d2hq9a1
not modelled
5.6
15
Fold: Split barrel-likeSuperfamily: FMN-binding split barrelFamily: PNP-oxidase like
41 d2fgca1
not modelled
5.6
27
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: IlvH-like
42 c3fhkF_
not modelled
5.5
54
PDB header: structural genomics, unknown functionChain: F: PDB Molecule: upf0403 protein yphp;PDBTitle: crystal structure of apc1446, b.subtilis yphp disulfide isomerase
43 d1rp1a2
not modelled
5.5
27
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Pancreatic lipase, N-terminal domain
44 c1zwvA_
not modelled
5.4
19
PDB header: transferaseChain: A: PDB Molecule: lipoamide acyltransferase component of branched-PDBTitle: solution structure of the subunit binding domain (hbsbd) of2 the human mitochondrial branched-chain alpha-ketoacid3 dehydrogenase
45 c3u0iA_
not modelled
5.3
12
PDB header: unknown functionChain: A: PDB Molecule: probable fad-binding, putative uncharacterized protein;PDBTitle: crystal structure of a probable fad-binding, putative uncharacterized2 protein from brucella melitensis
46 c1v06A_
not modelled
5.3
22
PDB header: dna-binding proteinChain: A: PDB Molecule: hmg box-containing protein 1;PDBTitle: axh domain of the transcription factor hbp1 from m.musculus
47 c3j39k_
not modelled
5.2
33
PDB header: ribosomeChain: K: PDB Molecule: 60s ribosomal protein l12;PDBTitle: structure of the d. melanogaster 60s ribosomal proteins
48 c2penE_
not modelled
5.2
20
PDB header: chaperoneChain: E: PDB Molecule: orf134;PDBTitle: crystal structure of rbcx, crystal form i
49 d1bala_
not modelled
5.1
38
Fold: Peripheral subunit-binding domain of 2-oxo acid dehydrogenase complexSuperfamily: Peripheral subunit-binding domain of 2-oxo acid dehydrogenase complexFamily: Peripheral subunit-binding domain of 2-oxo acid dehydrogenase complex
50 c1w4kA_
not modelled
5.1
22
PDB header: transferaseChain: A: PDB Molecule: pyruvate dehydrogenase e2;PDBTitle: peripheral-subunit binding domains from mesophilic,2 thermophilic, and hyperthermophilic bacteria fold by3 ultrafast, apparently two-state transitions
51 c3izcn_
not modelled
5.0
47
PDB header: ribosomeChain: N: PDB Molecule: 60s ribosomal protein rpl14 (l14e);PDBTitle: localization of the large subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome