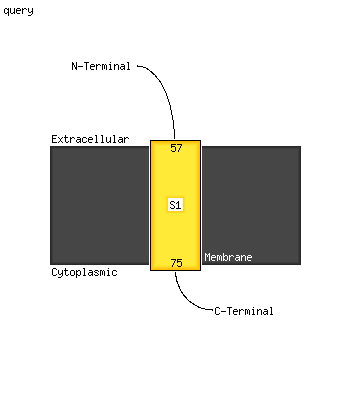
1 c1wolA_
87.7
11
PDB header: unknown functionChain: A: PDB Molecule: 122aa long conserved hypothetical protein;PDBTitle: crystal structure of st0689, an archaeal hepn homologue
2 c2hsbA_
77.4
11
PDB header: unknown functionChain: A: PDB Molecule: hypothetical upf0332 protein af0298;PDBTitle: crystal structure of a hepn domain containing protein (af_0298) from2 archaeoglobus fulgidus at 1.95 a resolution
3 d1ufba_
71.8
19
Fold: Four-helical up-and-down bundleSuperfamily: Nucleotidyltransferase substrate binding subunit/domainFamily: HEPN domain
4 d2b7oa1
55.6
17
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class-II DAHP synthetase
5 c2kncA_
35.7
10
PDB header: cell adhesionChain: A: PDB Molecule: integrin alpha-iib;PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
6 c5v2sA_
32.8
14
PDB header: viral proteinChain: A: PDB Molecule: envelope glycoprotein b;PDBTitle: crystal structure of glycoprotein b from herpes simplex virus type i
7 c1r7gA_
30.5
26
PDB header: membrane proteinChain: A: PDB Molecule: genome polyprotein;PDBTitle: nmr structure of the membrane anchor domain (1-31) of the2 nonstructural protein 5a (ns5a) of hepatitis c virus3 (minimized average structure, sample in 100mm dpc)
8 c1r7cA_
30.5
26
PDB header: membrane proteinChain: A: PDB Molecule: genome polyprotein;PDBTitle: nmr structure of the membrane anchor domain (1-31) of the2 nonstructural protein 5a (ns5a) of hepatitis c virus3 (minimized average structure, sample in 50% tfe)
9 c5n9yB_
29.0
9
PDB header: membrane proteinChain: B: PDB Molecule: zinc transport protein zntb;PDBTitle: the full-length structure of zntb
10 c1v8cA_
28.5
31
PDB header: protein bindingChain: A: PDB Molecule: moad related protein;PDBTitle: crystal structure of moad related protein from thermus2 thermophilus hb8
11 c3j0cG_
22.6
24
PDB header: virusChain: G: PDB Molecule: e1 envelope glycoprotein;PDBTitle: models of e1, e2 and cp of venezuelan equine encephalitis virus tc-832 strain restrained by a near atomic resolution cryo-em map
12 c2ks1A_
21.6
15
PDB header: transferaseChain: A: PDB Molecule: receptor tyrosine-protein kinase erbb-2;PDBTitle: heterodimeric association of transmembrane domains of erbb1 and erbb22 receptors enabling kinase activation
13 c2jwaA_
21.6
15
PDB header: transferaseChain: A: PDB Molecule: receptor tyrosine-protein kinase erbb-2;PDBTitle: erbb2 transmembrane segment dimer spatial structure
14 c5kwpA_
21.0
33
PDB header: de novo proteinChain: A: PDB Molecule: designed peptide nc_eeh_d2;PDBTitle: nmr solution structure of designed peptide nc_eeh_d2
15 c2k1aA_
20.4
11
PDB header: cell adhesionChain: A: PDB Molecule: integrin alpha-iib;PDBTitle: bicelle-embedded integrin alpha(iib) transmembrane segment
16 c5a5tC_
20.4
15
PDB header: hydrolaseChain: C: PDB Molecule: eukaryotic translation initiation factor 3 subunit c;PDBTitle: structure of mammalian eif3 in the context of the 43s preinitiation2 complex
17 c4u1cC_
19.6
17
PDB header: translationChain: C: PDB Molecule: eukaryotic translation initiation factor 3 subunit c;PDBTitle: crystal structure of the eif3a/eif3c pci-domain heterodimer
18 c4ev6E_
17.1
9
PDB header: metal transportChain: E: PDB Molecule: magnesium transport protein cora;PDBTitle: the complete structure of cora magnesium transporter from2 methanocaldococcus jannaschii
19 c3j7jC_
17.0
15
PDB header: translationChain: C: PDB Molecule: eukaryotic translation initiation factor 3 subunit c;PDBTitle: model of the human eif3 pci-mpn octamer docked into the 43s-hcv ires2 em map
20 c3j8bC_
17.0
15
PDB header: translationChain: C: PDB Molecule: eukaryotic translation initiation factor 3 subunit c;PDBTitle: model of the human eif3 pci-mpn octamer docked into the 43s-hcv ires2 em map
21 c3j8cC_
not modelled
17.0
15
PDB header: translationChain: C: PDB Molecule: eukaryotic translation initiation factor 3 subunit c;PDBTitle: model of the human eif3 pci-mpn octamer docked into the 43s em map
22 c3j7kC_
not modelled
17.0
15
PDB header: translationChain: C: PDB Molecule: eukaryotic translation initiation factor 3 subunit c;PDBTitle: model of the human eif3 pci-mpn octamer docked into the 43s em map
23 c2l8sA_
not modelled
15.9
8
PDB header: cell adhesionChain: A: PDB Molecule: integrin alpha-1;PDBTitle: solution nmr structure of transmembrane and cytosolic regions of2 integrin alpha1 in detergent micelles
24 c3dpiA_
not modelled
13.3
13
PDB header: ligaseChain: A: PDB Molecule: nad+ synthetase;PDBTitle: crystal structure of nad+ synthetase from burkholderia pseudomallei
25 c3o10D_
not modelled
13.1
13
PDB header: chaperoneChain: D: PDB Molecule: sacsin;PDBTitle: crystal structure of the hepn domain from human sacsin
26 d1wpga4
not modelled
12.2
13
Fold: Calcium ATPase, transmembrane domain MSuperfamily: Calcium ATPase, transmembrane domain MFamily: Calcium ATPase, transmembrane domain M
27 c2fzvC_
not modelled
11.9
16
PDB header: structural genomics, unknown functionChain: C: PDB Molecule: putative arsenical resistance protein;PDBTitle: crystal structure of an apo form of a flavin-binding protein from2 shigella flexneri
28 c3mk7F_
not modelled
11.7
23
PDB header: oxidoreductaseChain: F: PDB Molecule: cytochrome c oxidase, cbb3-type, subunit p;PDBTitle: the structure of cbb3 cytochrome oxidase
29 c2n2aA_
not modelled
10.2
15
PDB header: membrane proteinChain: A: PDB Molecule: receptor tyrosine-protein kinase erbb-2;PDBTitle: spatial structure of her2/erbb2 dimeric transmembrane domain in the2 presence of cytoplasmic juxtamembrane domains
30 c6mctH_
not modelled
10.0
5
PDB header: de novo proteinChain: H: PDB Molecule: mini-evgl membrane protein;PDBTitle: a designed pentameric membrane protein stabilized by van der waals2 interaction
31 c6mctO_
not modelled
10.0
5
PDB header: de novo proteinChain: O: PDB Molecule: mini-evgl membrane protein;PDBTitle: a designed pentameric membrane protein stabilized by van der waals2 interaction
32 c6mctC_
not modelled
10.0
5
PDB header: de novo proteinChain: C: PDB Molecule: mini-evgl membrane protein;PDBTitle: a designed pentameric membrane protein stabilized by van der waals2 interaction
33 c6mctJ_
not modelled
10.0
5
PDB header: de novo proteinChain: J: PDB Molecule: mini-evgl membrane protein;PDBTitle: a designed pentameric membrane protein stabilized by van der waals2 interaction
34 c6mctI_
not modelled
10.0
5
PDB header: de novo proteinChain: I: PDB Molecule: mini-evgl membrane protein;PDBTitle: a designed pentameric membrane protein stabilized by van der waals2 interaction
35 c6mctE_
not modelled
10.0
5
PDB header: de novo proteinChain: E: PDB Molecule: mini-evgl membrane protein;PDBTitle: a designed pentameric membrane protein stabilized by van der waals2 interaction
36 c6mctD_
not modelled
10.0
5
PDB header: de novo proteinChain: D: PDB Molecule: mini-evgl membrane protein;PDBTitle: a designed pentameric membrane protein stabilized by van der waals2 interaction
37 c6mctG_
not modelled
10.0
5
PDB header: de novo proteinChain: G: PDB Molecule: mini-evgl membrane protein;PDBTitle: a designed pentameric membrane protein stabilized by van der waals2 interaction
38 c6mctM_
not modelled
10.0
5
PDB header: de novo proteinChain: M: PDB Molecule: mini-evgl membrane protein;PDBTitle: a designed pentameric membrane protein stabilized by van der waals2 interaction
39 c6mctB_
not modelled
10.0
5
PDB header: de novo proteinChain: B: PDB Molecule: mini-evgl membrane protein;PDBTitle: a designed pentameric membrane protein stabilized by van der waals2 interaction
40 c6mctN_
not modelled
10.0
5
PDB header: de novo proteinChain: N: PDB Molecule: mini-evgl membrane protein;PDBTitle: a designed pentameric membrane protein stabilized by van der waals2 interaction
41 c6mctA_
not modelled
10.0
5
PDB header: de novo proteinChain: A: PDB Molecule: mini-evgl membrane protein;PDBTitle: a designed pentameric membrane protein stabilized by van der waals2 interaction
42 c6mctL_
not modelled
10.0
5
PDB header: de novo proteinChain: L: PDB Molecule: mini-evgl membrane protein;PDBTitle: a designed pentameric membrane protein stabilized by van der waals2 interaction
43 c6mctF_
not modelled
10.0
5
PDB header: de novo proteinChain: F: PDB Molecule: mini-evgl membrane protein;PDBTitle: a designed pentameric membrane protein stabilized by van der waals2 interaction
44 c6mq2D_
not modelled
10.0
5
PDB header: de novo proteinChain: D: PDB Molecule: mini-evgl membrane protein;PDBTitle: de novo design of membrane protein--mini-evgl membrane protein, c22212 form-2
45 c6mpwA_
not modelled
10.0
5
PDB header: de novo proteinChain: A: PDB Molecule: mini-evgl membrane protein;PDBTitle: de novo design of membrane protein--mini-evgl membrane protein, c22212 form-1
46 c6mctK_
not modelled
10.0
5
PDB header: de novo proteinChain: K: PDB Molecule: mini-evgl membrane protein;PDBTitle: a designed pentameric membrane protein stabilized by van der waals2 interaction
47 c2v1sD_
not modelled
9.9
25
PDB header: oxidoreductaseChain: D: PDB Molecule: mitochondrial import receptor subunit tom20 homolog;PDBTitle: crystal structure of rat tom20-aldh presequence complex
48 d1om2a_
not modelled
9.8
24
Fold: Open three-helical up-and-down bundleSuperfamily: Mitochondrial import receptor subunit Tom20Family: Mitochondrial import receptor subunit Tom20
49 c6mq2E_
not modelled
9.6
5
PDB header: de novo proteinChain: E: PDB Molecule: mini-evgl membrane protein;PDBTitle: de novo design of membrane protein--mini-evgl membrane protein, c22212 form-2
50 c6mpwE_
not modelled
9.6
5
PDB header: de novo proteinChain: E: PDB Molecule: mini-evgl membrane protein;PDBTitle: de novo design of membrane protein--mini-evgl membrane protein, c22212 form-1
51 c6mq2A_
not modelled
9.6
5
PDB header: de novo proteinChain: A: PDB Molecule: mini-evgl membrane protein;PDBTitle: de novo design of membrane protein--mini-evgl membrane protein, c22212 form-2
52 c6mpwB_
not modelled
9.6
5
PDB header: de novo proteinChain: B: PDB Molecule: mini-evgl membrane protein;PDBTitle: de novo design of membrane protein--mini-evgl membrane protein, c22212 form-1
53 c6mq2C_
not modelled
9.6
5
PDB header: de novo proteinChain: C: PDB Molecule: mini-evgl membrane protein;PDBTitle: de novo design of membrane protein--mini-evgl membrane protein, c22212 form-2
54 c6mpwC_
not modelled
9.6
5
PDB header: de novo proteinChain: C: PDB Molecule: mini-evgl membrane protein;PDBTitle: de novo design of membrane protein--mini-evgl membrane protein, c22212 form-1
55 c6mq2B_
not modelled
9.6
5
PDB header: de novo proteinChain: B: PDB Molecule: mini-evgl membrane protein;PDBTitle: de novo design of membrane protein--mini-evgl membrane protein, c22212 form-2
56 c6mpwD_
not modelled
9.6
5
PDB header: de novo proteinChain: D: PDB Molecule: mini-evgl membrane protein;PDBTitle: de novo design of membrane protein--mini-evgl membrane protein, c22212 form-1
57 d1o3ua_
not modelled
9.2
11
Fold: Four-helical up-and-down bundleSuperfamily: Nucleotidyltransferase substrate binding subunit/domainFamily: HEPN domain
58 c5t42A_
not modelled
9.2
12
PDB header: viral proteinChain: A: PDB Molecule: envelope glycoprotein;PDBTitle: structure of the ebola virus envelope protein mper/tm domain and its2 interaction with the fusion loop explains their fusion activity
59 c2axtH_
not modelled
9.0
15
PDB header: electron transportChain: H: PDB Molecule: photosystem ii reaction center h protein;PDBTitle: crystal structure of photosystem ii from thermosynechococcus elongatus
60 d2axth1
not modelled
9.0
15
Fold: Single transmembrane helixSuperfamily: Photosystem II 10 kDa phosphoprotein PsbHFamily: PsbH-like
61 c3urzB_
not modelled
8.4
10
PDB header: protein bindingChain: B: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a putative protein binding protein (bacova_03105)2 from bacteroides ovatus atcc 8483 at 2.19 a resolution
62 c5fshA_
not modelled
8.4
23
PDB header: hydrolaseChain: A: PDB Molecule: csm6;PDBTitle: crystal structure of thermus thermophilus csm6
63 c4nqfB_
not modelled
8.4
18
PDB header: transferaseChain: B: PDB Molecule: nucleotidyltransferase;PDBTitle: crystal structure of hepn domain protein
64 c6nk6B_
not modelled
7.9
20
PDB header: virus like particle/signaling proteinChain: B: PDB Molecule: e1 glycoprotein;PDBTitle: electron cryo-microscopy of chikungunya vlp in complex with mouse2 mxra8 receptor
65 c3q4gA_
not modelled
7.6
14
PDB header: ligaseChain: A: PDB Molecule: nh(3)-dependent nad(+) synthetase;PDBTitle: structure of nad synthetase from vibrio cholerae
66 c1zzaA_
not modelled
7.5
14
PDB header: membrane proteinChain: A: PDB Molecule: stannin;PDBTitle: solution nmr structure of the membrane protein stannin
67 c3u7rB_
not modelled
7.4
11
PDB header: oxidoreductaseChain: B: PDB Molecule: nadph-dependent fmn reductase;PDBTitle: ferb - flavoenzyme nad(p)h:(acceptor) oxidoreductase (ferb) from2 paracoccus denitrificans
68 c4rvqA_
not modelled
7.2
21
PDB header: protein bindingChain: A: PDB Molecule: pre-mrna splicing helicase-like protein;PDBTitle: pwi-like domain of chaetomium thermophilum brr2
69 d2f23a1
not modelled
7.0
15
Fold: Long alpha-hairpinSuperfamily: GreA transcript cleavage protein, N-terminal domainFamily: GreA transcript cleavage protein, N-terminal domain
70 c2xzeA_
not modelled
6.7
21
PDB header: hydrolase/protein transportChain: A: PDB Molecule: stam-binding protein;PDBTitle: structural basis for amsh-escrt-iii chmp3 interaction
71 c3hy5A_
not modelled
6.6
16
PDB header: transport proteinChain: A: PDB Molecule: retinaldehyde-binding protein 1;PDBTitle: crystal structure of cralbp
72 c4eu4A_
not modelled
6.6
15
PDB header: transferaseChain: A: PDB Molecule: succinyl-coa:acetate coenzyme a transferase;PDBTitle: succinyl-coa: acetate coa-transferase (aarch6) in complex with coa2 (hexagonal lattice)
73 c4jhjD_
not modelled
6.5
71
PDB header: translationChain: D: PDB Molecule: dead/h (asp-glu-ala-asp/his) box polypeptide 19 (dbp5PDBTitle: crystal structure of danio rerio slip1 in complex with dbp5
74 c4jhjC_
not modelled
6.5
71
PDB header: translationChain: C: PDB Molecule: dead/h (asp-glu-ala-asp/his) box polypeptide 19 (dbp5PDBTitle: crystal structure of danio rerio slip1 in complex with dbp5
75 c2nvvF_
not modelled
6.5
23
PDB header: hydrolaseChain: F: PDB Molecule: acetyl-coa hydrolase/transferase family protein;PDBTitle: crystal structure of the putative acetyl-coa hydrolase/transferase2 pg1013 from porphyromonas gingivalis, northeast structural genomics3 target pgr16.
76 c2vyiA_
not modelled
6.4
15
PDB header: chaperoneChain: A: PDB Molecule: sgta protein;PDBTitle: crystal structure of the tpr domain of human sgt
77 c1p58F_
not modelled
6.3
13
PDB header: virusChain: F: PDB Molecule: envelope protein m;PDBTitle: complex organization of dengue virus membrane proteins as revealed by2 9.5 angstrom cryo-em reconstruction
78 c1p58E_
not modelled
6.3
13
PDB header: virusChain: E: PDB Molecule: envelope protein m;PDBTitle: complex organization of dengue virus membrane proteins as revealed by2 9.5 angstrom cryo-em reconstruction
79 d1rkta2
not modelled
6.2
18
Fold: Tetracyclin repressor-like, C-terminal domainSuperfamily: Tetracyclin repressor-like, C-terminal domainFamily: Tetracyclin repressor-like, C-terminal domain
80 c3q49B_
not modelled
6.2
10
PDB header: ligase/chaperoneChain: B: PDB Molecule: stip1 homology and u box-containing protein 1;PDBTitle: crystal structure of the tpr domain of chip complexed with hsp70-c2 peptide
81 c6mx4J_
not modelled
6.0
25
PDB header: virusChain: J: PDB Molecule: e1;PDBTitle: cryoem structure of chimeric eastern equine encephalitis virus
82 c2l05A_
not modelled
5.9
15
PDB header: transferaseChain: A: PDB Molecule: serine/threonine-protein kinase b-raf;PDBTitle: solution nmr structure of the ras-binding domain of serine/threonine-2 protein kinase b-raf from homo sapiens, northeast structural genomics3 consortium target hr4694f
83 d1ecma_
not modelled
5.8
28
Fold: Chorismate mutase IISuperfamily: Chorismate mutase IIFamily: Dimeric chorismate mutase
84 c6agfB_
not modelled
5.8
2
PDB header: membrane proteinChain: B: PDB Molecule: sodium channel subunit beta-1;PDBTitle: structure of the human voltage-gated sodium channel nav1.4 in complex2 with beta1
85 c5hujB_
not modelled
5.8
13
PDB header: transferaseChain: B: PDB Molecule: nh(3)-dependent nad(+) synthetase;PDBTitle: crystal structure of nade from streptococcus pyogenes
86 c5yxiA_
not modelled
5.3
25
PDB header: de novo proteinChain: A: PDB Molecule: drafx6;PDBTitle: designed protein drafx6
87 c2q00B_
not modelled
5.3
19
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: orf c02003 protein;PDBTitle: crystal structure of the p95883_sulso protein from sulfolobus2 solfataricus. nesg target ssr10.
88 d1xrsa_
not modelled
5.2
17
Fold: TIM beta/alpha-barrelSuperfamily: Cobalamin (vitamin B12)-dependent enzymesFamily: D-lysine 5,6-aminomutase alpha subunit, KamD