1 c5fmrC_
98.8
13
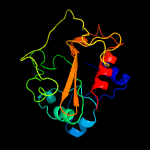
PDB header: transport proteinChain: C: PDB Molecule: intraflagellar transport protein component ift52;PDBTitle: crift52 n-terminal domain
2 c5fmsA_
98.6
13
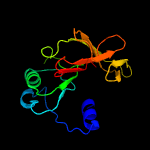
PDB header: transport proteinChain: A: PDB Molecule: intraflagellar transport protein 52 homolog;PDBTitle: mmift52 n-terminal domain
3 c5gslB_
96.1
15
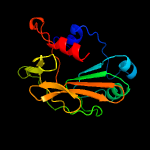
PDB header: hydrolaseChain: B: PDB Molecule: 778aa long hypothetical beta-galactosidase;PDBTitle: glycoside hydrolase a
4 c6eznG_
95.2
13
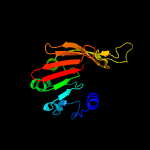
PDB header: membrane proteinChain: G: PDB Molecule: dolichyl-diphosphooligosaccharide--proteinPDBTitle: cryo-em structure of the yeast oligosaccharyltransferase (ost) complex
5 c5xb7E_
95.0
19
PDB header: hydrolaseChain: E: PDB Molecule: beta-galactosidase;PDBTitle: gh42 alpha-l-arabinopyranosidase from bifidobacterium animalis subsp.2 lactis bl-04
6 d1kwga3
94.9
16
Fold: Flavodoxin-likeSuperfamily: Class I glutamine amidotransferase-likeFamily: A4 beta-galactosidase middle domain
7 d2gk3a1
94.4
13
Fold: Flavodoxin-likeSuperfamily: Class I glutamine amidotransferase-likeFamily: STM3548-like
8 c1kwgA_
93.3
17
PDB header: hydrolaseChain: A: PDB Molecule: beta-galactosidase;PDBTitle: crystal structure of thermus thermophilus a4 beta-galactosidase
9 c5gsmB_
92.3
13
PDB header: hydrolaseChain: B: PDB Molecule: exo-beta-d-glucosaminidase;PDBTitle: glycoside hydrolase b with product
10 c4uzsB_
91.0
14
PDB header: hydrolaseChain: B: PDB Molecule: beta-galactosidase;PDBTitle: crystal structure of bifidobacterium bifidum beta-galactosidase
11 c4ojyA_
89.7
11
PDB header: hydrolaseChain: A: PDB Molecule: beta-galactosidase;PDBTitle: 3d structure of the e323a catalytic mutant of gan42b, a gh42 beta-2 galactosidase from g. stearothermophilus
12 c4uozC_
84.0
11
PDB header: hydrolaseChain: C: PDB Molecule: beta-galactosidase;PDBTitle: beta-(1,6)-galactosidase from bifidobacterium animalis subsp. lactis2 bl-04 nucleophile mutant e324a in complex with galactose
13 c5vymB_
83.5
17
PDB header: hydrolaseChain: B: PDB Molecule: beta-galactosidase bgab;PDBTitle: crystal structure of beta-galactosidase from bifidobacterium2 adolescentis
14 c3sozC_
83.5
11
PDB header: structural genomics, unknown functionChain: C: PDB Molecule: cytoplasmic protein stm1381;PDBTitle: cytoplasmic protein stm1381 from salmonella typhimurium lt2
15 d1x7da_
69.7
15
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Ornithine cyclodeaminase-like
16 c4e5vA_
66.6
13
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative thua-like protein;PDBTitle: crystal structure of a putative thua-like protein (parmer_02418) from2 parabacteroides merdae atcc 43184 at 1.75 a resolution
17 c2iv1J_
66.1
20
PDB header: lyaseChain: J: PDB Molecule: cyanate hydratase;PDBTitle: site directed mutagenesis of key residues involved in the catalytic2 mechanism of cyanase
18 c5e9aB_
64.2
12
PDB header: hydrolaseChain: B: PDB Molecule: beta-galactosidase;PDBTitle: crystal structure analysis of the cold-adamped beta-galactosidase from2 rahnella sp. r3
19 d1dwka1
58.2
19
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: Cyanase N-terminal domain
20 c3ttsD_
46.6
11
PDB header: hydrolaseChain: D: PDB Molecule: beta-galactosidase;PDBTitle: crystal structure of beta-galactosidase from bacillus circulans sp.2 alkalophilus
21 d1j6ua1
not modelled
45.0
19
Fold: MurCD N-terminal domainSuperfamily: MurCD N-terminal domainFamily: MurCD N-terminal domain
22 d1p3da1
not modelled
41.7
24
Fold: MurCD N-terminal domainSuperfamily: MurCD N-terminal domainFamily: MurCD N-terminal domain
23 c3mkzU_
not modelled
38.7
10
PDB header: dna-binding protein/dnaChain: U: PDB Molecule: protein sopb;PDBTitle: structure of sopb(155-272)-18mer complex, p21 form
24 c3mkyP_
not modelled
36.9
10
PDB header: dna binding protein/dnaChain: P: PDB Molecule: protein sopb;PDBTitle: structure of sopb(155-323)-18mer dna complex, i23 form
25 c5uk3J_
not modelled
35.7
14
PDB header: lyaseChain: J: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of cyanase from t. urticae
26 c1y9qA_
not modelled
33.9
20
PDB header: transcription regulatorChain: A: PDB Molecule: transcriptional regulator, hth_3 family;PDBTitle: crystal structure of hth_3 family transcriptional regulator2 from vibrio cholerae
27 c3rhtB_
not modelled
33.2
19
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: (gatase1)-like protein;PDBTitle: crystal structure of type 1 glutamine amidotransferase (gatase1)-like2 protein from planctomyces limnophilus
28 c3smaD_
not modelled
32.3
20
PDB header: transferaseChain: D: PDB Molecule: frbf;PDBTitle: a new n-acetyltransferase fold in the structure and mechanism of the2 phosphonate biosynthetic enzyme frbf
29 c6bc3A_
not modelled
31.0
20
PDB header: transferase/antibioticChain: A: PDB Molecule: aac 3-vi protein;PDBTitle: cryo x-ray structure of sisomicin bound aac-via
30 c6b9tH_
not modelled
30.8
21
PDB header: oxidoreductaseChain: H: PDB Molecule: methylphosphonate synthase;PDBTitle: crystal structure of mpns with substrate 2-hydroxyethylphosphonate (2-2 hep) and fe(ii) bound
31 c6mn5A_
not modelled
30.6
18
PDB header: transferase/antibioticChain: A: PDB Molecule: aminoglycoside n(3)-acetyltransferase, aac(3)-iva;PDBTitle: crystal structure of aminoglycoside acetyltransferase aac(3)-iva,2 h154a mutant, in complex with gentamicin c1a
32 d2nyga1
not modelled
29.5
15
Fold: TTHA0583/YokD-likeSuperfamily: TTHA0583/YokD-likeFamily: Aminoglycoside 3-N-acetyltransferase-like
33 c6b9rD_
not modelled
29.4
39
PDB header: oxidoreductaseChain: D: PDB Molecule: hydroxyethylphosphonate dioxygenase;PDBTitle: streptomyces albus hepd with substrate 2-hydroxyethylphosphonate (2-2 hep) and fe(ii) bound
34 d1t0ba_
not modelled
29.0
19
Fold: Flavodoxin-likeSuperfamily: Class I glutamine amidotransferase-likeFamily: ThuA-like
35 c4jqsC_
not modelled
26.9
6
PDB header: structural genomics, unknown functionChain: C: PDB Molecule: hypothetical protein;PDBTitle: crystal structure of a putative thua-like protein (bacuni_01602) from2 bacteroides uniformis atcc 8492 at 2.30 a resolution
36 c3e4fB_
not modelled
25.9
15
PDB header: transferaseChain: B: PDB Molecule: aminoglycoside n3-acetyltransferase;PDBTitle: crystal structure of ba2930- a putative aminoglycoside n3-2 acetyltransferase from bacillus anthracis
37 d1gr0a2
not modelled
25.3
25
Fold: FwdE/GAPDH domain-likeSuperfamily: Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domainFamily: Dihydrodipicolinate reductase-like
38 c6mb6A_
not modelled
25.2
18
PDB header: transferaseChain: A: PDB Molecule: aac(3)-iiib protein;PDBTitle: aac-iiib binary with coash
39 c4mcxE_
not modelled
23.7
24
PDB header: toxinChain: E: PDB Molecule: antidote protein;PDBTitle: p. vulgaris higba structure, crystal form 2
40 c1yi1A_
not modelled
23.3
33
PDB header: transferaseChain: A: PDB Molecule: acetolactate synthase;PDBTitle: crystal structure of arabidopsis thaliana acetohydroxyacid synthase in2 complex with a sulfonylurea herbicide, tribenuron methyl
41 c5gziB_
not modelled
23.3
22
PDB header: lyaseChain: B: PDB Molecule: lysine cyclodeaminase;PDBTitle: cyclodeaminase_pa
42 c4mp6A_
not modelled
22.8
3
PDB header: oxidoreductaseChain: A: PDB Molecule: putative ornithine cyclodeaminase;PDBTitle: staphyloferrin b precursor biosynthetic enzyme sbnb bound to citrate2 and nad+
43 c3cecA_
not modelled
22.5
16
PDB header: transcriptionChain: A: PDB Molecule: putative antidote protein of plasmid maintenance system;PDBTitle: crystal structure of a putative antidote protein of plasmid2 maintenance system (npun_f2943) from nostoc punctiforme pcc 73102 at3 1.60 a resolution
44 c3trbA_
not modelled
22.3
22
PDB header: dna binding proteinChain: A: PDB Molecule: virulence-associated protein i;PDBTitle: structure of an addiction module antidote protein of a higa (higa)2 family from coxiella burnetii
45 c2bnoA_
not modelled
21.3
21
PDB header: oxidoreductaseChain: A: PDB Molecule: epoxidase;PDBTitle: the structure of hydroxypropylphosphonic acid epoxidase from s.2 wedmorenis.
46 c6f8sA_
not modelled
21.3
19
PDB header: toxinChain: A: PDB Molecule: xre family transcriptional regulator;PDBTitle: toxin-antitoxin complex grata
47 c8jdwA_
not modelled
19.6
27
PDB header: transferaseChain: A: PDB Molecule: protein (l-arginine:glycine amidinotransferase);PDBTitle: crystal structure of human l-arginine:glycine amidinotransferase in2 complex with l-alanine
48 c1zzgB_
not modelled
19.4
21
PDB header: isomeraseChain: B: PDB Molecule: glucose-6-phosphate isomerase;PDBTitle: crystal structure of hypothetical protein tt0462 from thermus2 thermophilus hb8
49 d1ybha2
not modelled
19.2
35
Fold: Thiamin diphosphate-binding fold (THDP-binding)Superfamily: Thiamin diphosphate-binding fold (THDP-binding)Family: Pyruvate oxidase and decarboxylase Pyr module
50 d2icta1
not modelled
18.8
19
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: SinR domain-like
51 c1jdwA_
not modelled
18.5
27
PDB header: transferaseChain: A: PDB Molecule: l-arginine\:glycine amidinotransferase;PDBTitle: crystal structure and mechanism of l-arginine: glycine2 amidinotransferase: a mitochondrial enzyme involved in3 creatine biosynthesis
52 d1jdwa_
not modelled
18.5
27
Fold: Pentein, beta/alpha-propellerSuperfamily: PenteinFamily: Amidinotransferase
53 c5ht0B_
not modelled
17.3
20
PDB header: transferaseChain: B: PDB Molecule: aminoglycoside acetyltransferase hmb0005;PDBTitle: crystal structure of an antibiotic_nat family aminoglycoside2 acetyltransferase hmb0038 from an uncultured soil metagenomic sample3 in complex with coenzyme a
54 c2an1D_
not modelled
16.7
24
PDB header: transferaseChain: D: PDB Molecule: putative kinase;PDBTitle: structural genomics, the crystal structure of a putative kinase from2 salmonella typhimurim lt2
55 c1eysH_
not modelled
16.4
25
PDB header: electron transportChain: H: PDB Molecule: photosynthetic reaction center;PDBTitle: crystal structure of photosynthetic reaction center from a2 thermophilic bacterium, thermochromatium tepidum
56 c5wpiB_
not modelled
16.0
21
PDB header: transferaseChain: B: PDB Molecule: hsva;PDBTitle: the virulence-associated protein hsva from the fire blight pathogen2 erwinia amylovora is a polyamine amidinotransferase
57 c5d50I_
not modelled
15.4
19
PDB header: dna binding proteinChain: I: PDB Molecule: repressor;PDBTitle: crystal structure of rep-ant complex from salmonella-temperate phage
58 c1w8xP_
not modelled
14.7
14
PDB header: virusChain: P: PDB Molecule: protein p16;PDBTitle: structural analysis of prd1
59 c2dg2D_
not modelled
14.6
20
PDB header: protein bindingChain: D: PDB Molecule: apolipoprotein a-i binding protein;PDBTitle: crystal structure of mouse apolipoprotein a-i binding protein
60 c3mczB_
not modelled
14.3
10
PDB header: transferaseChain: B: PDB Molecule: o-methyltransferase;PDBTitle: the structure of an o-methyltransferase family protein from2 burkholderia thailandensis.
61 c3hdjA_
not modelled
13.5
12
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: probable ornithine cyclodeaminase;PDBTitle: the crystal structure of probable ornithine cyclodeaminase from2 bordetella pertussis tohama i
62 d1t6t1_
not modelled
13.1
15
Fold: Toprim domainSuperfamily: Toprim domainFamily: Toprim domain
63 c5emiA_
not modelled
12.9
28
PDB header: hydrolaseChain: A: PDB Molecule: cell wall hydrolase/autolysin;PDBTitle: n-acetylmuramoyl-l-alanine amidase amic2 of nostoc punctiforme
64 c2n1pA_
not modelled
12.7
20
PDB header: viral proteinChain: A: PDB Molecule: non-structural protein 5b, ns5b;PDBTitle: structure of the c-terminal membrane domain of hcv ns5b protein
65 c6ohiA_
not modelled
12.4
14
PDB header: biosynthetic proteinChain: A: PDB Molecule: debrominase bmp8;PDBTitle: crystal structure of the debrominase bmp8 (apo)
66 d2ihta2
not modelled
12.4
47
Fold: Thiamin diphosphate-binding fold (THDP-binding)Superfamily: Thiamin diphosphate-binding fold (THDP-binding)Family: Pyruvate oxidase and decarboxylase Pyr module
67 c2lonA_
not modelled
12.4
17
PDB header: membrane proteinChain: A: PDB Molecule: hig1 domain family member 1b;PDBTitle: backbone structure of human membrane protein higd1b
68 c3kgkA_
not modelled
12.2
18
PDB header: chaperoneChain: A: PDB Molecule: arsenical resistance operon trans-acting repressor arsd;PDBTitle: crystal structure of arsd
69 c2i99A_
not modelled
12.2
18
PDB header: oxidoreductaseChain: A: PDB Molecule: mu-crystallin homolog;PDBTitle: crystal structure of human mu_crystallin at 2.6 angstrom
70 d1u1ia2
not modelled
11.8
30
Fold: FwdE/GAPDH domain-likeSuperfamily: Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domainFamily: Dihydrodipicolinate reductase-like
71 c3eagA_
not modelled
11.1
29
PDB header: ligaseChain: A: PDB Molecule: udp-n-acetylmuramate:l-alanyl-gamma-d-glutamyl-meso-PDBTitle: the crystal structure of udp-n-acetylmuramate:l-alanyl-gamma-d-2 glutamyl-meso-diaminopimelate ligase (mpl) from neisseria3 meningitides
72 d2ji7a2
not modelled
10.7
25
Fold: Thiamin diphosphate-binding fold (THDP-binding)Superfamily: Thiamin diphosphate-binding fold (THDP-binding)Family: Pyruvate oxidase and decarboxylase Pyr module
73 c6cauA_
not modelled
10.6
17
PDB header: ligaseChain: A: PDB Molecule: udp-n-acetylmuramate--l-alanine ligase;PDBTitle: udp-n-acetylmuramate--alanine ligase from acinetobacter baumannii2 ab5075-uw with amppnp
74 c2ebyA_
not modelled
10.5
16
PDB header: transcriptionChain: A: PDB Molecule: putative hth-type transcriptional regulator ybaq;PDBTitle: crystal structure of a hypothetical protein from e. coli
75 c4zp3O_
not modelled
10.5
33
PDB header: signaling proteinChain: O: PDB Molecule: a-kinase anchor protein 7 isoforms alpha and beta;PDBTitle: akap18:pka-riialpha structure reveals crucial anchor points for2 recognition of regulatory subunits of pka
76 c4zp3R_
not modelled
10.5
33
PDB header: signaling proteinChain: R: PDB Molecule: a-kinase anchor protein 7 isoforms alpha and beta;PDBTitle: akap18:pka-riialpha structure reveals crucial anchor points for2 recognition of regulatory subunits of pka
77 c4zp3Q_
not modelled
10.5
33
PDB header: signaling proteinChain: Q: PDB Molecule: a-kinase anchor protein 7 isoforms alpha and beta;PDBTitle: akap18:pka-riialpha structure reveals crucial anchor points for2 recognition of regulatory subunits of pka
78 c1upaC_
not modelled
10.2
44
PDB header: synthaseChain: C: PDB Molecule: carboxyethylarginine synthase;PDBTitle: carboxyethylarginine synthase from streptomyces2 clavuligerus (semet structure)
79 c4v0bA_
not modelled
10.1
25
PDB header: hydrolaseChain: A: PDB Molecule: atp-dependent zinc metalloprotease ftsh;PDBTitle: escherichia coli ftsh hexameric n-domain
80 d1vjpa2
not modelled
10.1
10
Fold: FwdE/GAPDH domain-likeSuperfamily: Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domainFamily: Dihydrodipicolinate reductase-like
81 d1p1ja2
not modelled
9.6
30
Fold: FwdE/GAPDH domain-likeSuperfamily: Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domainFamily: Dihydrodipicolinate reductase-like
82 c2l9uA_
not modelled
9.1
21
PDB header: membrane proteinChain: A: PDB Molecule: receptor tyrosine-protein kinase erbb-3;PDBTitle: spatial structure of dimeric erbb3 transmembrane domain
83 c2l9uB_
not modelled
9.1
21
PDB header: membrane proteinChain: B: PDB Molecule: receptor tyrosine-protein kinase erbb-3;PDBTitle: spatial structure of dimeric erbb3 transmembrane domain
84 c2f00A_
not modelled
9.0
19
PDB header: ligaseChain: A: PDB Molecule: udp-n-acetylmuramate--l-alanine ligase;PDBTitle: escherichia coli murc
85 c4binA_
not modelled
9.0
14
PDB header: hydrolaseChain: A: PDB Molecule: n-acetylmuramoyl-l-alanine amidase amic;PDBTitle: crystal structure of the e. coli n-acetylmuramoyl-l-alanine amidase2 amic
86 c3eusB_
not modelled
8.6
17
PDB header: dna binding proteinChain: B: PDB Molecule: dna-binding protein;PDBTitle: the crystal structure of the dna binding protein from silicibacter2 pomeroyi
87 c1ta9A_
not modelled
8.3
5
PDB header: oxidoreductaseChain: A: PDB Molecule: glycerol dehydrogenase;PDBTitle: crystal structure of glycerol dehydrogenase from schizosaccharomyces2 pombe
88 c2lowA_
not modelled
8.1
13
PDB header: membrane proteinChain: A: PDB Molecule: apelin receptor;PDBTitle: solution structure of ar55 in 50% hfip
89 d1rioa_
not modelled
8.0
17
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: Phage repressors
90 c5zc2B_
not modelled
8.0
10
PDB header: flavoproteinChain: B: PDB Molecule: p-hydroxyphenylacetate 3-hydroxylase, reductase component;PDBTitle: acinetobacter baumannii p-hydroxyphenylacetate 3-hydroxylase (hpah),2 reductase component (c1)
91 d1vkoa2
not modelled
7.8
20
Fold: FwdE/GAPDH domain-likeSuperfamily: Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domainFamily: Dihydrodipicolinate reductase-like
92 c3ktbD_
not modelled
7.6
18
PDB header: transcription regulatorChain: D: PDB Molecule: arsenical resistance operon trans-acting repressor;PDBTitle: crystal structure of arsenical resistance operon trans-acting2 repressor from bacteroides vulgatus atcc 8482
93 c2xgjA_
not modelled
7.6
14
PDB header: hydrolase/rnaChain: A: PDB Molecule: atp-dependent rna helicase dob1;PDBTitle: structure of mtr4, a dexh helicase involved in nuclear rna2 processing and surveillance
94 d2jgra1
not modelled
7.5
20
Fold: NAD kinase/diacylglycerol kinase-likeSuperfamily: NAD kinase/diacylglycerol kinase-likeFamily: Diacylglycerol kinase-like
95 d1ozha2
not modelled
7.3
24
Fold: Thiamin diphosphate-binding fold (THDP-binding)Superfamily: Thiamin diphosphate-binding fold (THDP-binding)Family: Pyruvate oxidase and decarboxylase Pyr module
96 d1adra_
not modelled
7.2
13
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: Phage repressors
97 c6iegA_
not modelled
6.9
21
PDB header: rna binding proteinChain: A: PDB Molecule: exosome rna helicase mtr4;PDBTitle: crystal structure of human mtr4
98 d2ff4a2
not modelled
6.8
19
Fold: alpha-alpha superhelixSuperfamily: TPR-likeFamily: BTAD-like
99 c2lcoA_
not modelled
6.7
24
PDB header: membrane proteinChain: A: PDB Molecule: walp19-p8 peptide;PDBTitle: 1h and 15n assignments of walp19-p8 peptide in sds micelles