| 1 | c2d4wA_
|
|
 |
100.0 |
62 |
PDB header:transferase
Chain: A: PDB Molecule:glycerol kinase;
PDBTitle: crystal structure of glycerol kinase from cellulomonas sp.2 nt3060
|
|
|
|
| 2 | c3flcX_
|
|
 |
100.0 |
49 |
PDB header:transferase
Chain: X: PDB Molecule:glycerol kinase;
PDBTitle: crystal structure of the his-tagged h232r mutant of glycerol kinase2 from enterococcus casseliflavus with glycerol
|
|
|
|
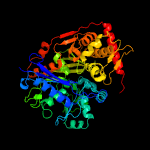
| 3 | c3g25B_
|
|
 |
100.0 |
49 |
PDB header:transferase
Chain: B: PDB Molecule:glycerol kinase;
PDBTitle: 1.9 angstrom crystal structure of glycerol kinase (glpk) from2 staphylococcus aureus in complex with glycerol.
|
|
|
|
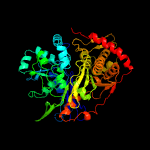
| 4 | c1glbG_
|
|
 |
100.0 |
51 |
PDB header:phosphotransferase
Chain: G: PDB Molecule:glycerol kinase;
PDBTitle: structure of the regulatory complex of escherichia coli iiiglc with2 glycerol kinase
|
|
|
|
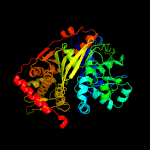
| 5 | c2zf5O_
|
|
 |
100.0 |
50 |
PDB header:transferase
Chain: O: PDB Molecule:glycerol kinase;
PDBTitle: crystal structure of highly thermostable glycerol kinase from a2 hyperthermophilic archaeon
|
|
|
|
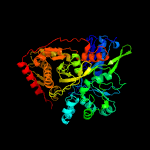
| 6 | c3ezwD_
|
|
 |
100.0 |
52 |
PDB header:transferase
Chain: D: PDB Molecule:glycerol kinase;
PDBTitle: crystal structure of a hyperactive escherichia coli glycerol kinase2 mutant gly230 --> asp obtained using microfluidic crystallization3 devices
|
|
|
|
| 7 | c2dpnB_
|
|
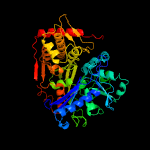 |
100.0 |
52 |
PDB header:transferase
Chain: B: PDB Molecule:glycerol kinase;
PDBTitle: crystal structure of the glycerol kinase from thermus2 thermophilus hb8
|
|
|
|
| 8 | c3wxiB_
|
|
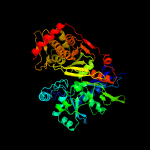 |
100.0 |
46 |
PDB header:transferase
Chain: B: PDB Molecule:glycerol kinase;
PDBTitle: crystal structure of trypanosoma brucei gambiense glycerol kinase2 (ligand-free form)
|
|
|
|
| 9 | c2w40C_
|
|
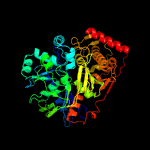 |
100.0 |
41 |
PDB header:transferase
Chain: C: PDB Molecule:glycerol kinase, putative;
PDBTitle: crystal structure of plasmodium falciparum glycerol kinase2 with bound glycerol
|
|
|
|
| 10 | c4e1jA_
|
|
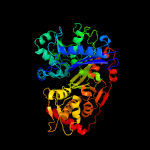 |
100.0 |
47 |
PDB header:transferase
Chain: A: PDB Molecule:glycerol kinase;
PDBTitle: crystal structure of glycerol kinase in complex with glycerol from2 sinorhizobium meliloti 1021
|
|
|
|
| 11 | c3gbtA_
|
|
 |
100.0 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:gluconate kinase;
PDBTitle: crystal structure of gluconate kinase from lactobacillus acidophilus
|
|
|
|
| 12 | c1xupO_
|
|
 |
100.0 |
50 |
PDB header:transferase
Chain: O: PDB Molecule:glycerol kinase;
PDBTitle: enterococcus casseliflavus glycerol kinase complexed with glycerol
|
|
|
|
| 13 | c5vm1A_
|
|
 |
100.0 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:xylulokinase;
PDBTitle: crystal structure of a xyloylose kinase from brucella ovis
|
|
|
|
| 14 | c3ifrB_
|
|
 |
100.0 |
24 |
PDB header:transferase
Chain: B: PDB Molecule:carbohydrate kinase, fggy;
PDBTitle: the crystal structure of xylulose kinase from rhodospirillum rubrum
|
|
|
|
| 15 | c3hz6A_
|
|
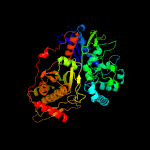 |
100.0 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:xylulokinase;
PDBTitle: crystal structure of xylulokinase from chromobacterium violaceum
|
|
|
|
| 16 | c2nlxA_
|
|
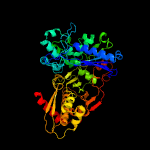 |
100.0 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:xylulose kinase;
PDBTitle: crystal structure of the apo e. coli xylulose kinase
|
|
|
|
| 17 | c3jvpA_
|
|
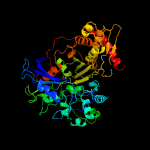 |
100.0 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:ribulokinase;
PDBTitle: crystal structure of ribulokinase from bacillus halodurans
|
|
|
|
| 18 | c3gg4B_
|
|
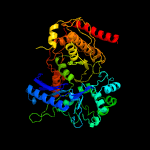 |
100.0 |
20 |
PDB header:transferase
Chain: B: PDB Molecule:glycerol kinase;
PDBTitle: the crystal structure of glycerol kinase from yersinia2 pseudotuberculosis
|
|
|
|
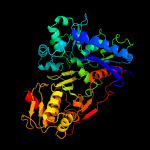
| 19 | c4bc2A_
|
|
 |
100.0 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:xylulose kinase;
PDBTitle: crystal structure of human d-xylulokinase in complex with d-2 xylulose and adenosine diphosphate
|
|
|
|
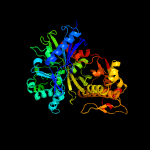
| 20 | c5ya2A_
|
|
 |
100.0 |
24 |
PDB header:structural protein
Chain: A: PDB Molecule:autoinducer-2 kinase;
PDBTitle: crystal structure of lsrk-hpr complex with adp
|
|
|
|
| 21 | c2cgkB_ |
|
not modelled |
100.0 |
17 |
PDB header:transferase
Chain: B: PDB Molecule:l-rhamnulose kinase;
PDBTitle: crystal structure of l-rhamnulose kinase from escherichia coli in an2 open uncomplexed conformation.
|
|
|
| 22 | c4c23A_ |
|
not modelled |
100.0 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:l-fuculose kinase fuck;
PDBTitle: l-fuculose kinase
|
|
|
| 23 | c3i8bA_ |
|
not modelled |
100.0 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:xylulose kinase;
PDBTitle: the crystal structure of xylulose kinase from2 bifidobacterium adolescentis
|
|
|
| 24 | c5hv7A_ |
|
not modelled |
100.0 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:probable sugar kinase;
PDBTitle: putative sugar kinases from synechococcus elongatus pcc7942 in complex2 with d-ribulose
|
|
|
| 25 | c5htxA_ |
|
not modelled |
100.0 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:putative xylulose kinase;
PDBTitle: putative sugar kinases from arabidopsis thaliana in complex with adp
|
|
|
| 26 | c3h6eB_ |
|
not modelled |
100.0 |
16 |
PDB header:transferase
Chain: B: PDB Molecule:carbohydrate kinase, fggy;
PDBTitle: the crystal structure of a carbohydrate kinase from novosphingobium2 aromaticivorans
|
|
|
| 27 | d2p3ra1 |
|
not modelled |
100.0 |
51 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:Glycerol kinase |
|
|
| 28 | d1r59o1 |
|
not modelled |
100.0 |
48 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:Glycerol kinase |
|
|
| 29 | d2p3ra2 |
|
not modelled |
100.0 |
51 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:Glycerol kinase |
|
|
| 30 | d1r59o2 |
|
not modelled |
100.0 |
53 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:Glycerol kinase |
|
|
| 31 | c3h1qB_ |
|
not modelled |
99.4 |
17 |
PDB header:structural protein
Chain: B: PDB Molecule:ethanolamine utilization protein eutj;
PDBTitle: crystal structure of ethanolamine utilization protein eutj from2 carboxydothermus hydrogenoformans
|
|
|
| 32 | d1huxa_ |
|
not modelled |
99.3 |
21 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:BadF/BadG/BcrA/BcrD-like |
|
|
| 33 | c4ehtA_ |
|
not modelled |
99.3 |
19 |
PDB header:electron transport
Chain: A: PDB Molecule:activator of 2-hydroxyisocaproyl-coa dehydratase;
PDBTitle: activator of the 2-hydroxyisocaproyl-coa dehydratase from clostridium2 difficile with bound adp
|
|
|
| 34 | c2ap1A_ |
|
not modelled |
99.0 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:putative regulator protein;
PDBTitle: crystal structure of the putative regulatory protein
|
|
|
| 35 | d1zc6a1 |
|
not modelled |
98.9 |
21 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:BadF/BadG/BcrA/BcrD-like |
|
|
| 36 | c2e2pA_ |
|
not modelled |
98.9 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:hexokinase;
PDBTitle: crystal structure of sulfolobus tokodaii hexokinase in2 complex with adp
|
|
|
| 37 | d2ewsa1 |
|
not modelled |
98.9 |
18 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:Fumble-like |
|
|
| 38 | c2ivoC_ |
|
not modelled |
98.7 |
12 |
PDB header:hydrolase
Chain: C: PDB Molecule:up1;
PDBTitle: structure of up1 protein
|
|
|
| 39 | c3enoB_ |
|
not modelled |
98.7 |
15 |
PDB header:hydrolase/unknown function
Chain: B: PDB Molecule:putative o-sialoglycoprotein endopeptidase;
PDBTitle: crystal structure of pyrococcus furiosus pcc1 in complex2 with thermoplasma acidophilum kae1
|
|
|
| 40 | d1z05a3 |
|
not modelled |
98.6 |
8 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:ROK |
|
|
| 41 | d2hoea3 |
|
not modelled |
98.5 |
7 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:ROK |
|
|
| 42 | c3eo3B_ |
|
not modelled |
98.5 |
21 |
PDB header:isomerase, transferase
Chain: B: PDB Molecule:bifunctional udp-n-acetylglucosamine 2-epimerase/n-
PDBTitle: crystal structure of the n-acetylmannosamine kinase domain of human2 gne protein
|
|
|
| 43 | c1zc6A_ |
|
not modelled |
98.5 |
19 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:probable n-acetylglucosamine kinase;
PDBTitle: crystal structure of putative n-acetylglucosamine kinase from2 chromobacterium violaceum. northeast structural genomics target3 cvr23.
|
|
|
| 44 | c2qm1D_ |
|
not modelled |
98.4 |
17 |
PDB header:transferase
Chain: D: PDB Molecule:glucokinase;
PDBTitle: crystal structure of glucokinase from enterococcus faecalis
|
|
|
| 45 | d2ch5a2 |
|
not modelled |
98.4 |
14 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:BadF/BadG/BcrA/BcrD-like |
|
|
| 46 | c1z05A_ |
|
not modelled |
98.4 |
9 |
PDB header:transcription
Chain: A: PDB Molecule:transcriptional regulator, rok family;
PDBTitle: crystal structure of the rok family transcriptional regulator, homolog2 of e.coli mlc protein.
|
|
|
| 47 | d1z6ra2 |
|
not modelled |
98.4 |
11 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:ROK |
|
|
| 48 | c2ch5D_ |
|
not modelled |
98.3 |
12 |
PDB header:transferase
Chain: D: PDB Molecule:nagk protein;
PDBTitle: crystal structure of human n-acetylglucosamine kinase in2 complex with n-acetylglucosamine
|
|
|
| 49 | c1z6rC_ |
|
not modelled |
98.3 |
9 |
PDB header:transcription
Chain: C: PDB Molecule:mlc protein;
PDBTitle: crystal structure of mlc from escherichia coli
|
|
|
| 50 | c4htlA_ |
|
not modelled |
98.2 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:beta-glucoside kinase;
PDBTitle: lmo2764 protein, a putative n-acetylmannosamine kinase, from listeria2 monocytogenes
|
|
|
| 51 | c4db3A_ |
|
not modelled |
98.2 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:n-acetyl-d-glucosamine kinase;
PDBTitle: 1.95 angstrom resolution crystal structure of n-acetyl-d-glucosamine2 kinase from vibrio vulnificus.
|
|
|
| 52 | c3zyyX_ |
|
not modelled |
98.2 |
18 |
PDB header:iron-sulfur-binding protein
Chain: X: PDB Molecule:iron-sulfur cluster binding protein;
PDBTitle: reductive activator for corrinoid,iron-sulfur protein
|
|
|
| 53 | c5f7rA_ |
|
not modelled |
98.2 |
10 |
PDB header:transcription
Chain: A: PDB Molecule:lmo0178 protein;
PDBTitle: rok repressor lmo0178 from listeria monocytogenes bound to inducer
|
|
|
| 54 | c3htvA_ |
|
not modelled |
98.2 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:d-allose kinase;
PDBTitle: crystal structure of d-allose kinase (np_418508.1) from escherichia2 coli k12 at 1.95 a resolution
|
|
|
| 55 | c3zyyY_ |
|
not modelled |
98.2 |
20 |
PDB header:iron-sulfur-binding protein
Chain: Y: PDB Molecule:iron-sulfur cluster binding protein;
PDBTitle: reductive activator for corrinoid,iron-sulfur protein
|
|
|
| 56 | c2hoeA_ |
|
not modelled |
98.1 |
7 |
PDB header:transferase
Chain: A: PDB Molecule:n-acetylglucosamine kinase;
PDBTitle: crystal structure of n-acetylglucosamine kinase (tm1224) from2 thermotoga maritima at 2.46 a resolution
|
|
|
| 57 | d1woqa1 |
|
not modelled |
98.0 |
20 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:ROK |
|
|
| 58 | c1dkgD_ |
|
not modelled |
97.9 |
16 |
PDB header:complex (hsp24/hsp70)
Chain: D: PDB Molecule:molecular chaperone dnak;
PDBTitle: crystal structure of the nucleotide exchange factor grpe2 bound to the atpase domain of the molecular chaperone dnak
|
|
|
| 59 | d2ap1a2 |
|
not modelled |
97.9 |
13 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:ROK |
|
|
| 60 | c5nckA_ |
|
not modelled |
97.9 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:n-acetylmannosamine kinase;
PDBTitle: the crystal structure of n-acetylmannosamine kinase in fusobacterium2 nucleatum
|
|
|
| 61 | c4yh5B_ |
|
not modelled |
97.8 |
13 |
PDB header:transferase
Chain: B: PDB Molecule:levoglucosan kinase;
PDBTitle: lipomyces starkeyi levoglucosan kinase bound to adp and manganese
|
|
|
| 62 | d1q18a1 |
|
not modelled |
97.8 |
19 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:Glucokinase |
|
|
| 63 | c3r8eA_ |
|
not modelled |
97.8 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:hypothetical sugar kinase;
PDBTitle: crystal structure of a putative sugar kinase (chu_1875) from cytophaga2 hutchinsonii atcc 33406 at 1.65 a resolution
|
|
|
| 64 | d1sz2a1 |
|
not modelled |
97.8 |
16 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:Glucokinase |
|
|
| 65 | c3vgkB_ |
|
not modelled |
97.8 |
17 |
PDB header:transferase
Chain: B: PDB Molecule:glucokinase;
PDBTitle: crystal structure of a rok family glucokinase from streptomyces2 griseus
|
|
|
| 66 | d1dkgd2 |
|
not modelled |
97.7 |
16 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:Actin/HSP70 |
|
|
| 67 | c4kboA_ |
|
not modelled |
97.7 |
17 |
PDB header:signaling protein
Chain: A: PDB Molecule:stress-70 protein, mitochondrial;
PDBTitle: crystal structure of the human mortalin (grp75) atpase domain in the2 apo form
|
|
|
| 68 | c5f7pA_ |
|
not modelled |
97.7 |
13 |
PDB header:transcription
Chain: A: PDB Molecule:lmo0178 protein;
PDBTitle: rok repressor lmo0178 from listeria monocytogenes
|
|
|
| 69 | c3mcpA_ |
|
not modelled |
97.7 |
9 |
PDB header:transferase
Chain: A: PDB Molecule:glucokinase;
PDBTitle: crystal structure of glucokinase (bdi_1628) from parabacteroides2 distasonis atcc 8503 at 3.00 a resolution
|
|
|
| 70 | d2aa4a1 |
|
not modelled |
97.7 |
13 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:ROK |
|
|
| 71 | c1e4gT_ |
|
not modelled |
97.6 |
9 |
PDB header:bacterial cell division
Chain: T: PDB Molecule:cell division protein ftsa;
PDBTitle: ftsa (atp-bound form) from thermotoga maritima
|
|
|
| 72 | c2v7zA_ |
|
not modelled |
97.6 |
14 |
PDB header:chaperone
Chain: A: PDB Molecule:heat shock cognate 71 kda protein;
PDBTitle: crystal structure of the 70-kda heat shock cognate protein2 from rattus norvegicus in post-atp hydrolysis state
|
|
|
| 73 | d2gupa1 |
|
not modelled |
97.6 |
23 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:ROK |
|
|
| 74 | c3d2fC_ |
|
not modelled |
97.6 |
13 |
PDB header:chaperone
Chain: C: PDB Molecule:heat shock protein homolog sse1;
PDBTitle: crystal structure of a complex of sse1p and hsp70
|
|
|
| 75 | c2aa4B_ |
|
not modelled |
97.6 |
19 |
PDB header:transferase
Chain: B: PDB Molecule:putative n-acetylmannosamine kinase;
PDBTitle: crystal structure of escherichia coli putative n-2 acetylmannosamine kinase, new york structural genomics3 consortium
|
|
|
| 76 | c2v7yA_ |
|
not modelled |
97.6 |
18 |
PDB header:chaperone
Chain: A: PDB Molecule:chaperone protein dnak;
PDBTitle: crystal structure of the molecular chaperone dnak from2 geobacillus kaustophilus hta426 in post-atp hydrolysis3 state
|
|
|
| 77 | c6gfaA_ |
|
not modelled |
97.6 |
18 |
PDB header:chaperone
Chain: A: PDB Molecule:heat shock protein 105 kda;
PDBTitle: structure of nucleotide binding domain of hsp110, atp and mg2+2 complexed
|
|
|
| 78 | c4gniA_ |
|
not modelled |
97.6 |
16 |
PDB header:chaperone
Chain: A: PDB Molecule:putative heat shock protein;
PDBTitle: structure of the ssz1 atpase bound to atp and magnesium
|
|
|
| 79 | d1e4ft1 |
|
not modelled |
97.6 |
11 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:Actin/HSP70 |
|
|
| 80 | c6da0A_ |
|
not modelled |
97.6 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:glucokinase;
PDBTitle: crystal structure of glucokinase (nfhk) from naegleria fowleri
|
|
|
| 81 | c1xc3A_ |
|
not modelled |
97.5 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:putative fructokinase;
PDBTitle: structure of a putative fructokinase from bacillus subtilis
|
|
|
| 82 | c1hpmA_ |
|
not modelled |
97.5 |
13 |
PDB header:hydrolase (acting on acid anhydrides)
Chain: A: PDB Molecule:44k atpase fragment (n-terminal) of 7o kd heat-
PDBTitle: how potassium affects the activity of the molecular2 chaperone hsc70. ii. potassium binds specifically in the3 atpase active site
|
|
|
| 83 | c4ijaA_ |
|
not modelled |
97.5 |
7 |
PDB header:protein binding
Chain: A: PDB Molecule:xylr protein;
PDBTitle: structure of s. aureus methicillin resistance factor mecr2
|
|
|
| 84 | c3wqtB_ |
|
not modelled |
97.5 |
16 |
PDB header:structural genomics
Chain: B: PDB Molecule:cell division protein ftsa;
PDBTitle: staphylococcus aureus ftsa complexed with amppnp
|
|
|
| 85 | d1jcea2 |
|
not modelled |
97.5 |
15 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:Actin/HSP70 |
|
|
| 86 | d2e8aa2 |
|
not modelled |
97.4 |
17 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:Actin/HSP70 |
|
|
| 87 | c3vewA_ |
|
not modelled |
97.4 |
25 |
PDB header:transferase
Chain: A: PDB Molecule:o-carbamoyltransferase tobz;
PDBTitle: crystal structure of the o-carbamoyltransferase tobz in complex with2 adp
|
|
|
| 88 | d1bupa2 |
|
not modelled |
97.4 |
14 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:Actin/HSP70 |
|
|
| 89 | c5mb9B_ |
|
not modelled |
97.4 |
19 |
PDB header:chaperone
Chain: B: PDB Molecule:putative heat shock protein;
PDBTitle: crystal structure of the eukaryotic ribosome associated complex (rac),2 a unique hsp70/hsp40 pair
|
|
|
| 90 | c3zeuE_ |
|
not modelled |
97.4 |
18 |
PDB header:hydrolase
Chain: E: PDB Molecule:probable trna threonylcarbamoyladenosine biosynthesis
PDBTitle: structure of a salmonella typhimurium ygjd-yeaz heterodimer bound to2 atpgammas
|
|
|
| 91 | c3vovC_ |
|
not modelled |
97.4 |
15 |
PDB header:transferase
Chain: C: PDB Molecule:glucokinase;
PDBTitle: crystal structure of rok hexokinase from thermus thermophilus
|
|
|
| 92 | c5e84B_ |
|
not modelled |
97.4 |
18 |
PDB header:chaperone
Chain: B: PDB Molecule:78 kda glucose-regulated protein;
PDBTitle: atp-bound state of bip
|
|
|
| 93 | c4bgaB_ |
|
not modelled |
97.4 |
16 |
PDB header:transferase
Chain: B: PDB Molecule:predicted molecular chaperone distantly related to
PDBTitle: nucleotide-bound open form of a putative sugar kinase2 mk0840 from methanopyrus kandleri
|
|
|
| 94 | d1xc3a1 |
|
not modelled |
97.3 |
12 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:ROK |
|
|
| 95 | c5tkyA_ |
|
not modelled |
97.3 |
16 |
PDB header:chaperone
Chain: A: PDB Molecule:putative uncharacterized protein;
PDBTitle: crystal structure of the co-translational hsp70 chaperone ssb in the2 atp-bound, open conformation
|
|
|
| 96 | c2q2rA_ |
|
not modelled |
97.3 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:glucokinase 1, putative;
PDBTitle: trypanosoma cruzi glucokinase in complex with beta-d-glucose and adp
|
|
|
| 97 | c4czeA_ |
|
not modelled |
97.3 |
16 |
PDB header:structural protein
Chain: A: PDB Molecule:rod shape-determining protein mreb;
PDBTitle: c. crescentus mreb, double filament, empty
|
|
|
| 98 | c3vthA_ |
|
not modelled |
97.3 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:hydrogenase maturation factor;
PDBTitle: crystal structure of full-length hypf in the phosphate- and2 nucleotide-bound form
|
|
|
| 99 | c6ediA_ |
|
not modelled |
97.3 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:glucokinase;
PDBTitle: crystal structure of leishmania braziliensis glucokinase
|
|
|
| 100 | c3c7nB_ |
|
not modelled |
97.3 |
15 |
PDB header:chaperone/chaperone
Chain: B: PDB Molecule:heat shock cognate;
PDBTitle: structure of the hsp110:hsc70 nucleotide exchange complex
|
|
|
| 101 | c2gupA_ |
|
not modelled |
97.2 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:rok family protein;
PDBTitle: structural genomics, the crystal structure of a rok family protein2 from streptococcus pneumoniae tigr4 in complex with sucrose
|
|
|
| 102 | c6gwjK_ |
|
not modelled |
97.2 |
17 |
PDB header:rna binding protein
Chain: K: PDB Molecule:probable trna n6-adenosine threonylcarbamoyltransferase;
PDBTitle: protein complex
|
|
|
| 103 | c3cqyA_ |
|
not modelled |
97.2 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:anhydro-n-acetylmuramic acid kinase;
PDBTitle: crystal structure of a functionally unknown protein (so_1313) from2 shewanella oneidensis mr-1
|
|
|
| 104 | c1woqB_ |
|
not modelled |
97.2 |
20 |
PDB header:transferase
Chain: B: PDB Molecule:inorganic polyphosphate/atp-glucomannokinase;
PDBTitle: crystal structure of inorganic polyphosphate/atp-glucomannokinase from2 arthrobacter sp. strain km at 1.8 a resolution
|
|
|
| 105 | c3vthB_ |
|
not modelled |
97.2 |
14 |
PDB header:transferase
Chain: B: PDB Molecule:hydrogenase maturation factor;
PDBTitle: crystal structure of full-length hypf in the phosphate- and2 nucleotide-bound form
|
|
|
| 106 | c3iucC_ |
|
not modelled |
97.2 |
18 |
PDB header:chaperone
Chain: C: PDB Molecule:heat shock 70kda protein 5 (glucose-regulated
PDBTitle: crystal structure of the human 70kda heat shock protein 52 (bip/grp78) atpase domain in complex with adp
|
|
|
| 107 | c1jcgA_ |
|
not modelled |
97.2 |
14 |
PDB header:structural protein
Chain: A: PDB Molecule:rod shape-determining protein mreb;
PDBTitle: mreb from thermotoga maritima, amppnp
|
|
|
| 108 | c4rtfD_ |
|
not modelled |
97.1 |
17 |
PDB header:chaperone
Chain: D: PDB Molecule:chaperone protein dnak;
PDBTitle: crystal structure of molecular chaperone dnak from mycobacterium2 tuberculosis h37rv
|
|
|
| 109 | c5obuA_ |
|
not modelled |
97.1 |
18 |
PDB header:chaperone
Chain: A: PDB Molecule:chaperone protein dnak;
PDBTitle: mycoplasma genitalium dnak deletion mutant lacking sbdalpha in complex2 with amppnp.
|
|
|
| 110 | c3vpzA_ |
|
not modelled |
97.1 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:glucokinase;
PDBTitle: crystal structure of glucokinase from antarctic psychrotroph at 1.69a
|
|
|
| 111 | d1bg3a3 |
|
not modelled |
97.0 |
17 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:Hexokinase |
|
|
| 112 | c2khoA_ |
|
not modelled |
97.0 |
21 |
PDB header:chaperone
Chain: A: PDB Molecule:heat shock protein 70;
PDBTitle: nmr-rdc / xray structure of e. coli hsp70 (dnak) chaperone (1-605)2 complexed with adp and substrate
|
|
|
| 113 | c6fpeG_ |
|
not modelled |
96.9 |
12 |
PDB header:rna binding protein
Chain: G: PDB Molecule:trna n6-adenosine threonylcarbamoyltransferase;
PDBTitle: bacterial protein complex
|
|
|
| 114 | d1bdga1 |
|
not modelled |
96.9 |
17 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:Hexokinase |
|
|
| 115 | c4j8fA_ |
|
not modelled |
96.8 |
18 |
PDB header:chaperone
Chain: A: PDB Molecule:heat shock 70 kda protein 1a/1b, hsc70-interacting protein;
PDBTitle: crystal structure of a fusion protein containing the nbd of hsp70 and2 the middle domain of hip
|
|
|
| 116 | c5eoxB_ |
|
not modelled |
96.8 |
17 |
PDB header:peptide binding protein
Chain: B: PDB Molecule:type 4 fimbrial biogenesis protein pilm;
PDBTitle: pseudomonas aeruginosa pilm bound to adp
|
|
|
| 117 | c1bdgA_ |
|
not modelled |
96.6 |
17 |
PDB header:hexokinase
Chain: A: PDB Molecule:hexokinase;
PDBTitle: hexokinase from schistosoma mansoni complexed with glucose
|
|
|
| 118 | d1v4sa1 |
|
not modelled |
96.6 |
14 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:Hexokinase |
|
|
| 119 | d1e4ft2 |
|
not modelled |
96.5 |
15 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:Actin/HSP70 |
|
|
| 120 | c3qbwA_ |
|
not modelled |
96.5 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:anhydro-n-acetylmuramic acid kinase;
PDBTitle: crystal structure of pseudomonas aeruginosa 1,6-anhydro-n-2 actetylmuramic acid kinase (anmk) bound to adenosine diphosphate
|
|
|