1 c4ymkA_
82.0
15
PDB header: oxidoreductaseChain: A: PDB Molecule: acyl-coa desaturase 1;PDBTitle: crystal structure of stearoyl-coenzyme a desaturase 1
2 c2m0qA_
69.5
23
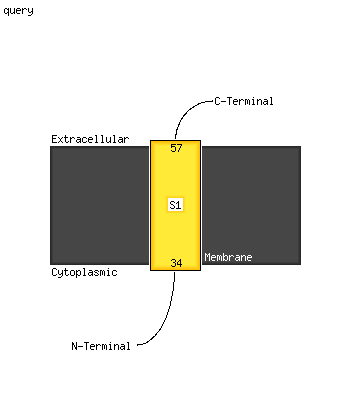
PDB header: membrane proteinChain: A: PDB Molecule: potassium voltage-gated channel subfamily e member 2;PDBTitle: solution nmr analysis of intact kcne2 in detergent micelles2 demonstrate a straight transmembrane helix
3 c2k21A_
54.5
27
PDB header: membrane proteinChain: A: PDB Molecule: potassium voltage-gated channel subfamily ePDBTitle: nmr structure of human kcne1 in lmpg micelles at ph 6.0 and2 40 degree c
4 c6a69B_
48.4
29
PDB header: structural proteinChain: B: PDB Molecule: neuroplastin;PDBTitle: cryo-em structure of a p-type atpase
5 c6a0aA_
38.7
48
PDB header: structural proteinChain: A: PDB Molecule: collagen type iii peptide;PDBTitle: structure of a triple-helix region of human collagen type iii
6 c1x1kF_
36.8
68
PDB header: structural proteinChain: F: PDB Molecule: host-guest peptide (pro-pro-gly)4-(pro-allohyp-PDBTitle: host-guest peptide (pro-pro-gly)4-(pro-allohyp-gly)-(pro-2 pro-gly)4
7 c6a0aC_
34.0
52
PDB header: structural proteinChain: C: PDB Molecule: collagen type iii peptide;PDBTitle: structure of a triple-helix region of human collagen type iii
8 c6a0aB_
34.0
52
PDB header: structural proteinChain: B: PDB Molecule: collagen type iii peptide;PDBTitle: structure of a triple-helix region of human collagen type iii
9 c2jo1A_
29.2
23
PDB header: hydrolase regulatorChain: A: PDB Molecule: phospholemman;PDBTitle: structure of the na,k-atpase regulatory protein fxyd1 in2 micelles
10 c6nhyA_
29.0
38
PDB header: immune systemChain: A: PDB Molecule: tumor necrosis factor receptor superfamily member 10b;PDBTitle: structure of the transmembrane domain of the death receptor 5 mutant2 (g217y) - trimer only
11 c6nhyB_
29.0
38
PDB header: immune systemChain: B: PDB Molecule: tumor necrosis factor receptor superfamily member 10b;PDBTitle: structure of the transmembrane domain of the death receptor 5 mutant2 (g217y) - trimer only
12 c6nhyC_
29.0
38
PDB header: immune systemChain: C: PDB Molecule: tumor necrosis factor receptor superfamily member 10b;PDBTitle: structure of the transmembrane domain of the death receptor 5 mutant2 (g217y) - trimer only
13 c2mkvA_
28.7
23
PDB header: transport proteinChain: A: PDB Molecule: sodium/potassium-transporting atpase subunit gamma;PDBTitle: structure of the na,k-atpase regulatory protein fxyd2b in micelles
14 c3jcuj_
28.5
47
PDB header: membrane proteinChain: J: PDB Molecule: photosystem ii reaction center protein j;PDBTitle: cryo-em structure of spinach psii-lhcii supercomplex at 3.2 angstrom2 resolution
15 c6a0cB_
28.1
48
PDB header: structural proteinChain: B: PDB Molecule: collagen type iii peptide;PDBTitle: structure of a triple-helix region of human collagen type iii
16 c6hwhB_
26.2
13
PDB header: electron transportChain: B: PDB Molecule: ubiquinol-cytochrome c reductase iron-sulfur subunit;PDBTitle: structure of a functional obligate respiratory supercomplex from2 mycobacterium smegmatis
17 c5xnmj_
25.9
44
PDB header: membrane proteinChain: J: PDB Molecule: photosystem ii reaction center protein j;PDBTitle: structure of unstacked c2s2m2-type psii-lhcii supercomplex from pisum2 sativum
18 c4zyoA_
25.1
23
PDB header: oxidoreductaseChain: A: PDB Molecule: acyl-coa desaturase;PDBTitle: crystal structure of human integral membrane stearoyl-coa desaturase2 with substrate
19 c2k1vB_
25.0
54
PDB header: hormoneChain: B: PDB Molecule: relaxin-3;PDBTitle: r3/i5 relaxin chimera
20 c2k1aA_
24.6
13
PDB header: cell adhesionChain: A: PDB Molecule: integrin alpha-iib;PDBTitle: bicelle-embedded integrin alpha(iib) transmembrane segment
21 c2ndjA_
not modelled
24.5
25
PDB header: membrane proteinChain: A: PDB Molecule: potassium voltage-gated channel subfamily e member 3;PDBTitle: structural basis for kcne3 and estrogen modulation of the kcnq12 channel
22 c2jp3A_
not modelled
24.4
21
PDB header: transcriptionChain: A: PDB Molecule: fxyd domain-containing ion transport regulator 4;PDBTitle: solution structure of the human fxyd4 (chif) protein in sds2 micelles
23 c5oqtC_
not modelled
23.6
45
PDB header: transport proteinChain: C: PDB Molecule: uncharacterized protein ynem;PDBTitle: crystal structure of a bacterial cationic amino acid transporter (cat)2 homologue
24 c6f34C_
not modelled
23.6
45
PDB header: membrane proteinChain: C: PDB Molecule: mgts;PDBTitle: crystal structure of a bacterial cationic amino acid transporter (cat)2 homologue bound to arginine.
25 c6a0cC_
not modelled
23.5
52
PDB header: structural proteinChain: C: PDB Molecule: collagen type iii peptide;PDBTitle: structure of a triple-helix region of human collagen type iii
26 c6a0cA_
not modelled
23.5
52
PDB header: structural proteinChain: A: PDB Molecule: collagen type iii peptide;PDBTitle: structure of a triple-helix region of human collagen type iii
27 c3a0mD_
not modelled
19.9
64
PDB header: structural proteinChain: D: PDB Molecule: collagen-like peptide;PDBTitle: structure of (ppg)4-ovg-(ppg)4, monoclinic, twinned crystal
28 c2k9hA_
not modelled
19.9
67
PDB header: metal binding proteinChain: A: PDB Molecule: glycoprotein;PDBTitle: the hantavirus glycoprotein g1 tail contains a dual cchc-2 type classical zinc fingers
29 c6c14B_
not modelled
19.0
15
PDB header: membrane protein, metal transportChain: B: PDB Molecule: lhfpl tetraspan subfamily member 5 protein;PDBTitle: cryoem structure of mouse pcdh15-1ec-lhfpl5 complex
30 c3a0mB_
not modelled
18.8
65
PDB header: structural proteinChain: B: PDB Molecule: collagen-like peptide;PDBTitle: structure of (ppg)4-ovg-(ppg)4, monoclinic, twinned crystal
31 c4yzfA_
not modelled
18.5
19
PDB header: immune systemChain: A: PDB Molecule: band 3 anion transport protein;PDBTitle: crystal structure of the anion exchanger domain of human erythrocyte2 band 3
32 c3a0mE_
not modelled
17.9
58
PDB header: structural proteinChain: E: PDB Molecule: collagen-like peptide;PDBTitle: structure of (ppg)4-ovg-(ppg)4, monoclinic, twinned crystal
33 c3a0mC_
not modelled
17.9
64
PDB header: structural proteinChain: C: PDB Molecule: collagen-like peptide;PDBTitle: structure of (ppg)4-ovg-(ppg)4, monoclinic, twinned crystal
34 c2v53C_
not modelled
17.6
54
PDB header: cell adhesionChain: C: PDB Molecule: collagen alpha-1(iii) chain;PDBTitle: crystal structure of a sparc-collagen complex
35 c2v53D_
not modelled
17.6
54
PDB header: cell adhesionChain: D: PDB Molecule: collagen alpha-1(iii) chain;PDBTitle: crystal structure of a sparc-collagen complex
36 c2v53B_
not modelled
17.6
54
PDB header: cell adhesionChain: B: PDB Molecule: collagen alpha-1(iii) chain;PDBTitle: crystal structure of a sparc-collagen complex
37 c2k1kA_
not modelled
17.4
83
PDB header: signaling proteinChain: A: PDB Molecule: ephrin type-a receptor 1;PDBTitle: nmr structures of dimeric transmembrane domain of the2 receptor tyrosine kinase epha1 in lipid bicelles at ph 4.3
38 c2k1lB_
not modelled
17.4
83
PDB header: signaling proteinChain: B: PDB Molecule: ephrin type-a receptor 1;PDBTitle: nmr structures of dimeric transmembrane domain of the2 receptor tyrosine kinase epha1 in lipid bicelles at ph 6.3
39 c2k1lA_
not modelled
17.4
83
PDB header: signaling proteinChain: A: PDB Molecule: ephrin type-a receptor 1;PDBTitle: nmr structures of dimeric transmembrane domain of the2 receptor tyrosine kinase epha1 in lipid bicelles at ph 6.3
40 c2k1kB_
not modelled
17.4
83
PDB header: signaling proteinChain: B: PDB Molecule: ephrin type-a receptor 1;PDBTitle: nmr structures of dimeric transmembrane domain of the2 receptor tyrosine kinase epha1 in lipid bicelles at ph 4.3
41 c3ctwB_
not modelled
17.0
40
PDB header: protein bindingChain: B: PDB Molecule: rcda;PDBTitle: crystal structure of rcda from caulobacter crescentus cb15
42 d1ug0a_
not modelled
17.0
18
Fold: Surp module (SWAP domain)Superfamily: Surp module (SWAP domain)Family: Surp module (SWAP domain)
43 c3a1hA_
not modelled
16.2
58
PDB header: structural proteinChain: A: PDB Molecule: collagen-like peptide;PDBTitle: crystal structure analysis of the collagen-like peptide, (ppg)4-otg-2 (ppg)4
44 c2kncA_
not modelled
15.3
13
PDB header: cell adhesionChain: A: PDB Molecule: integrin alpha-iib;PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
45 c3a0mA_
not modelled
15.2
64
PDB header: structural proteinChain: A: PDB Molecule: collagen-like peptide;PDBTitle: structure of (ppg)4-ovg-(ppg)4, monoclinic, twinned crystal
46 c6caaA_
not modelled
14.8
12
PDB header: transport proteinChain: A: PDB Molecule: electrogenic sodium bicarbonate cotransporter 1;PDBTitle: cryoem structure of human slc4a4 sodium-coupled acid-base transporter2 nbce1
47 c3a1hF_
not modelled
14.7
65
PDB header: structural proteinChain: F: PDB Molecule: collagen-like peptide;PDBTitle: crystal structure analysis of the collagen-like peptide, (ppg)4-otg-2 (ppg)4
48 c3hcjB_
not modelled
14.5
40
PDB header: oxidoreductaseChain: B: PDB Molecule: peptide methionine sulfoxide reductase;PDBTitle: structure of msrb from xanthomonas campestris (oxidized2 form)
49 c6nhwC_
not modelled
13.6
54
PDB header: immune systemChain: C: PDB Molecule: tumor necrosis factor receptor superfamily member 10b;PDBTitle: structure of the transmembrane domain of the death receptor 5 - dimer2 of trimer
50 c6nhwE_
not modelled
13.6
54
PDB header: immune systemChain: E: PDB Molecule: tumor necrosis factor receptor superfamily member 10b;PDBTitle: structure of the transmembrane domain of the death receptor 5 - dimer2 of trimer
51 c6nhwF_
not modelled
13.6
54
PDB header: immune systemChain: F: PDB Molecule: tumor necrosis factor receptor superfamily member 10b;PDBTitle: structure of the transmembrane domain of the death receptor 5 - dimer2 of trimer
52 c6nhwB_
not modelled
13.6
54
PDB header: immune systemChain: B: PDB Molecule: tumor necrosis factor receptor superfamily member 10b;PDBTitle: structure of the transmembrane domain of the death receptor 5 - dimer2 of trimer
53 c3a1hE_
not modelled
13.5
64
PDB header: structural proteinChain: E: PDB Molecule: collagen-like peptide;PDBTitle: crystal structure analysis of the collagen-like peptide, (ppg)4-otg-2 (ppg)4
54 c3a1hC_
not modelled
13.5
64
PDB header: structural proteinChain: C: PDB Molecule: collagen-like peptide;PDBTitle: crystal structure analysis of the collagen-like peptide, (ppg)4-otg-2 (ppg)4
55 c3a1hD_
not modelled
13.5
64
PDB header: structural proteinChain: D: PDB Molecule: collagen-like peptide;PDBTitle: crystal structure analysis of the collagen-like peptide, (ppg)4-otg-2 (ppg)4
56 c3n23E_
not modelled
13.3
42
PDB header: hydrolaseChain: E: PDB Molecule: na+/k+ atpase gamma subunit transcript variant a;PDBTitle: crystal structure of the high affinity complex between ouabain and the2 e2p form of the sodium-potassium pump
57 d1x4oa1
not modelled
13.1
29
Fold: Surp module (SWAP domain)Superfamily: Surp module (SWAP domain)Family: Surp module (SWAP domain)
58 c4xydB_
not modelled
13.0
17
PDB header: oxidoreductaseChain: B: PDB Molecule: norc-like protein;PDBTitle: nitric oxide reductase from roseobacter denitrificans (rdnor)
59 d1qhda1
not modelled
12.8
83
Fold: A virus capsid protein alpha-helical domainSuperfamily: A virus capsid protein alpha-helical domainFamily: vp6, the major capsid protein of group A rotavirus
60 c6irtA_
not modelled
12.7
25
PDB header: membrane proteinChain: A: PDB Molecule: 4f2 cell-surface antigen heavy chain;PDBTitle: human lat1-4f2hc complex bound with bch
61 d1xcra1
not modelled
12.3
32
Fold: AF0104/ALDC/Ptd012-likeSuperfamily: AF0104/ALDC/Ptd012-likeFamily: PTD012-like
62 c3gcfC_
not modelled
12.2
62
PDB header: oxidoreductaseChain: C: PDB Molecule: terminal oxygenase component of carbazole 1,9a-PDBTitle: terminal oxygenase of carbazole 1,9a-dioxygenase from2 nocardioides aromaticivorans ic177
63 c3a1hB_
not modelled
12.2
58
PDB header: structural proteinChain: B: PDB Molecule: collagen-like peptide;PDBTitle: crystal structure analysis of the collagen-like peptide, (ppg)4-otg-2 (ppg)4
64 d2hlya1
not modelled
11.4
33
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: Atu2299-like
65 c5sv9B_
not modelled
11.4
9
PDB header: transport proteinChain: B: PDB Molecule: bor1p boron transporter;PDBTitle: structure of the slc4 transporter bor1p in an inward-facing2 conformation
66 d2axtj1
not modelled
11.4
50
Fold: Single transmembrane helixSuperfamily: Photosystem II reaction center protein J, PsbJFamily: PsbJ-like
67 c3a0hJ_
not modelled
11.4
50
PDB header: electron transportChain: J: PDB Molecule: photosystem ii reaction center protein j;PDBTitle: crystal structure of i-substituted photosystem ii complex
68 c6ae1B_
not modelled
11.1
50
PDB header: rna binding proteinChain: B: PDB Molecule: crispr-associated protein, tm1810 family;PDBTitle: crystal structure of csm2 of the type iii-a crispr-cas effector2 complex
69 c4bklG_
not modelled
11.1
100
PDB header: immune systemChain: G: PDB Molecule: j1 epitope;PDBTitle: crystal structure of the arthritogenic antibody m2139 (fab2 fragment) in complex with the triple-helical j1 peptide
70 c2e5zA_
not modelled
11.0
23
PDB header: rna binding proteinChain: A: PDB Molecule: splicing factor, arginine/serine-rich 8;PDBTitle: solution structure of the surp2 domain in splicing factor,2 arginine/serine-rich 8
71 c2hmnA_
not modelled
10.9
83
PDB header: oxidoreductaseChain: A: PDB Molecule: naphthalene 1,2-dioxygenase alpha subunit;PDBTitle: crystal structure of the naphthalene 1,2-dioxygenase f352v2 mutant bound to anthracene.
72 c1z01D_
not modelled
10.8
62
PDB header: oxidoreductaseChain: D: PDB Molecule: 2-oxo-1,2-dihydroquinoline 8-monooxygenase, oxygenasePDBTitle: 2-oxoquinoline 8-monooxygenase component: active site modulation by2 rieske-[2fe-2s] center oxidation/reduction
73 c3jcuI_
not modelled
10.6
29
PDB header: membrane proteinChain: I: PDB Molecule: protein photosystem ii reaction center protein i;PDBTitle: cryo-em structure of spinach psii-lhcii supercomplex at 3.2 angstrom2 resolution
74 c3kdpH_
not modelled
10.2
36
PDB header: hydrolaseChain: H: PDB Molecule: na+/k+ atpase gamma subunit transcript variant a;PDBTitle: crystal structure of the sodium-potassium pump
75 c3kdpG_
not modelled
10.2
36
PDB header: hydrolaseChain: G: PDB Molecule: na+/k+ atpase gamma subunit transcript variant a;PDBTitle: crystal structure of the sodium-potassium pump
76 c2hacB_
not modelled
10.2
33
PDB header: membrane proteinChain: B: PDB Molecule: t-cell surface glycoprotein cd3 zeta chain;PDBTitle: structure of zeta-zeta transmembrane dimer
77 c2hacA_
not modelled
10.2
33
PDB header: membrane proteinChain: A: PDB Molecule: t-cell surface glycoprotein cd3 zeta chain;PDBTitle: structure of zeta-zeta transmembrane dimer
78 c3nvnA_
not modelled
10.1
57
PDB header: viral protein/signaling proteinChain: A: PDB Molecule: evm139;PDBTitle: molecular mechanism of guidance cue recognition
79 c3gzkA_
not modelled
9.9
28
PDB header: hydrolaseChain: A: PDB Molecule: cellulase;PDBTitle: structure of a. acidocaldarius cellulase cela
80 d2axti1
not modelled
9.8
35
Fold: Single transmembrane helixSuperfamily: Photosystem II reaction center protein I, PsbIFamily: PsbI-like
81 c3a0hi_
not modelled
9.8
35
PDB header: electron transportChain: I: PDB Molecule: photosystem ii reaction center protein i;PDBTitle: crystal structure of i-substituted photosystem ii complex
82 c6c3rB_
not modelled
9.8
63
PDB header: viral proteinChain: B: PDB Molecule: cricket paralysis virus 1a protein;PDBTitle: cricket paralysis virus rnai suppressor protein crpv-1a
83 c4hqjG_
not modelled
9.8
36
PDB header: hydrolase/transport proteinChain: G: PDB Molecule: na+/k+ atpase gamma subunit transcript variant a;PDBTitle: crystal structure of na+,k+-atpase in the na+-bound state
84 c5vnyA_
not modelled
9.7
71
PDB header: endocytosis, protein bindingChain: A: PDB Molecule: lethal (2) giant discs 1, isoform b;PDBTitle: crystal structure of dm14-3 domain of lgd
85 c1wqlA_
not modelled
9.1
62
PDB header: oxidoreductaseChain: A: PDB Molecule: iron-sulfur protein large subunit of cumene dioxygenase;PDBTitle: cumene dioxygenase (cuma1a2) from pseudomonas fluorescens ip01
86 c4hqjE_
not modelled
8.9
36
PDB header: hydrolase/transport proteinChain: E: PDB Molecule: na+/k+ atpase gamma subunit transcript variant a;PDBTitle: crystal structure of na+,k+-atpase in the na+-bound state
87 c3abnC_
not modelled
8.9
65
PDB header: structural proteinChain: C: PDB Molecule: collagen-like peptide;PDBTitle: crystal structure of (pro-pro-gly)4-hyp-asp-gly-(pro-pro-gly)4 at 1.022 a
88 c3admC_
not modelled
8.8
64
PDB header: structural proteinChain: C: PDB Molecule: collagen-like peptide;PDBTitle: crystal structure of (pro-pro-gly)4-hyp-ser-gly-(pro-pro-gly)4
89 c2de7B_
not modelled
8.7
54
PDB header: oxidoreductaseChain: B: PDB Molecule: terminal oxygenase component of carbazole;PDBTitle: the substrate-bound complex between oxygenase and2 ferredoxin in carbazole 1,9a-dioxygenase
90 c3admD_
not modelled
8.6
64
PDB header: structural proteinChain: D: PDB Molecule: collagen-like peptide;PDBTitle: crystal structure of (pro-pro-gly)4-hyp-ser-gly-(pro-pro-gly)4
91 c3a0aF_
not modelled
8.6
65
PDB header: structural proteinChain: F: PDB Molecule: collagen-like peptide;PDBTitle: structure of (ppg)4-opg-(ppg)4, monoclinic, twinned crystal
92 c4bmtB_
not modelled
8.5
37
PDB header: oxidoreductaseChain: B: PDB Molecule: ribonucleoside-diphosphate reductase subunit beta;PDBTitle: crystal structure of ribonucleotide reductase di-iron nrdf2 from bacillus cereus
93 c3a0aA_
not modelled
8.5
58
PDB header: structural proteinChain: A: PDB Molecule: collagen-like peptide;PDBTitle: structure of (ppg)4-opg-(ppg)4, monoclinic, twinned crystal
94 c2elpA_
not modelled
8.4
63
PDB header: transcriptionChain: A: PDB Molecule: zinc finger protein 406;PDBTitle: solution structure of the 13th c2h2 zinc finger of human2 zinc finger protein 406
95 c3a08F_
not modelled
8.2
65
PDB header: structural proteinChain: F: PDB Molecule: collagen-like peptide;PDBTitle: structure of (ppg)4-oog-(ppg)4, monoclinic, twinned crystal
96 c3abnB_
not modelled
8.2
64
PDB header: structural proteinChain: B: PDB Molecule: collagen-like peptide;PDBTitle: crystal structure of (pro-pro-gly)4-hyp-asp-gly-(pro-pro-gly)4 at 1.022 a
97 c3eamB_
not modelled
7.8
23
PDB header: membrane protein, transport proteinChain: B: PDB Molecule: glr4197 protein;PDBTitle: an open-pore structure of a bacterial pentameric ligand-2 gated ion channel
98 c3a08E_
not modelled
7.8
58
PDB header: structural proteinChain: E: PDB Molecule: collagen-like peptide;PDBTitle: structure of (ppg)4-oog-(ppg)4, monoclinic, twinned crystal
99 c2kogA_
not modelled
7.6
12
PDB header: membrane proteinChain: A: PDB Molecule: vesicle-associated membrane protein 2;PDBTitle: lipid-bound synaptobrevin solution nmr structure