| 1 |
|
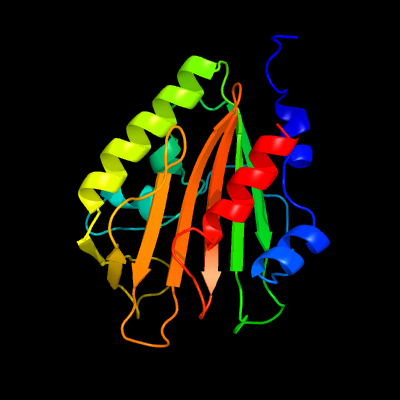
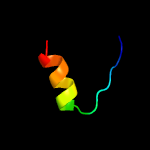
PDB 4esq chain A
Region: 24 - 213
Aligned: 188
Modelled: 190
Confidence: 100.0%
Identity: 20%
PDB header:transferase
Chain: A: PDB Molecule:serine/threonine protein kinase;
PDBTitle: crystal structure of the extracellular domain of pknh from2 mycobacterium tuberculosis
Phyre2
| 2 |
|
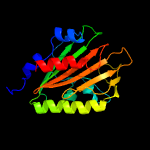
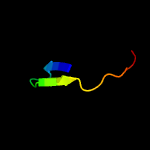
PDB 3lo3 chain E
Region: 110 - 126
Aligned: 17
Modelled: 17
Confidence: 39.7%
Identity: 24%
PDB header:structure genomics, unknown function
Chain: E: PDB Molecule:uncharacterized conserved protein;
PDBTitle: the crystal structure of a conserved functionally unknown protein from2 colwellia psychrerythraea 34h.
Phyre2
| 3 |
|
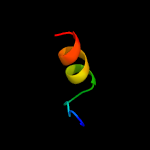
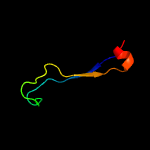
PDB 2fiu chain A domain 1
Region: 110 - 126
Aligned: 17
Modelled: 17
Confidence: 39.2%
Identity: 24%
Fold: Ferredoxin-like
Superfamily: Dimeric alpha+beta barrel
Family: Atu0297-like
Phyre2
| 4 |
|
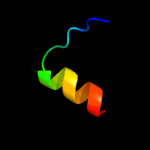
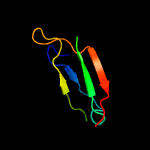
PDB 2mhe chain B
Region: 85 - 141
Aligned: 51
Modelled: 57
Confidence: 28.2%
Identity: 24%
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized protein;
PDBTitle: nmr structure of the protein np_419126.1 from caulobacter crescentus
Phyre2
| 5 |
|
PDB 2b3g chain B
Region: 27 - 43
Aligned: 17
Modelled: 17
Confidence: 26.5%
Identity: 12%
PDB header:replication
Chain: B: PDB Molecule:cellular tumor antigen p53;
PDBTitle: p53n (fragment 33-60) bound to rpa70n
Phyre2
| 6 |
|
PDB 3i08 chain D
Region: 114 - 130
Aligned: 17
Modelled: 17
Confidence: 23.9%
Identity: 12%
PDB header:signaling protein
Chain: D: PDB Molecule:neurogenic locus notch homolog protein 1;
PDBTitle: crystal structure of the s1-cleaved notch1 negative2 regulatory region (nrr)
Phyre2
| 7 |
|
PDB 5hb8 chain B
Region: 98 - 127
Aligned: 27
Modelled: 30
Confidence: 13.1%
Identity: 37%
PDB header:transport protein
Chain: B: PDB Molecule:nucleoporin nup53;
PDBTitle: crystal structure of chaetomium thermophilum nup53 rrm (space group2 p3121)
Phyre2
| 8 |
|
PDB 2l14 chain B
Region: 27 - 43
Aligned: 17
Modelled: 17
Confidence: 12.5%
Identity: 12%
PDB header:protein binding
Chain: B: PDB Molecule:cellular tumor antigen p53;
PDBTitle: structure of cbp nuclear coactivator binding domain in complex with2 p53 tad
Phyre2
| 9 |
|
PDB 4dwl chain A
Region: 24 - 49
Aligned: 26
Modelled: 26
Confidence: 11.4%
Identity: 31%
PDB header:nucleic acid binding protein
Chain: A: PDB Molecule:bbp7;
PDBTitle: avd molecule from bordetella bacteriophage dgr
Phyre2
| 10 |
|
PDB 2ly4 chain B
Region: 27 - 43
Aligned: 17
Modelled: 17
Confidence: 11.3%
Identity: 12%
PDB header:nuclear protein/antitumour protein
Chain: B: PDB Molecule:cellular tumor antigen p53;
PDBTitle: hmgb1-facilitated p53 dna binding occurs via hmg-box/p532 transactivation domain interaction and is regulated by the acidic3 tail
Phyre2
| 11 |
|
PDB 5uz5 chain C
Region: 109 - 127
Aligned: 19
Modelled: 19
Confidence: 8.9%
Identity: 32%
PDB header:nuclear protein/rna
Chain: C: PDB Molecule:u1 small nuclear ribonucleoprotein a,tap tag;
PDBTitle: s. cerevisiae u1 snrnp
Phyre2
| 12 |
|
PDB 2pp6 chain A domain 1
Region: 78 - 98
Aligned: 21
Modelled: 21
Confidence: 7.8%
Identity: 10%
Fold: Phage tail proteins
Superfamily: Phage tail proteins
Family: gpFII-like
Phyre2
| 13 |
|
PDB 3dca chain C
Region: 85 - 126
Aligned: 42
Modelled: 38
Confidence: 7.2%
Identity: 7%
PDB header:structural genomics, unknown function
Chain: C: PDB Molecule:rpa0582;
PDBTitle: crystal structure of the rpa0582- protein of unknown2 function from rhodopseudomonas palustris- a structural3 genomics target
Phyre2
| 14 |
|
PDB 2ker chain A
Region: 149 - 195
Aligned: 47
Modelled: 47
Confidence: 6.6%
Identity: 15%
PDB header:hydrolase inhibitor
Chain: A: PDB Molecule:alpha-amylase inhibitor z-2685;
PDBTitle: alpha-amylase inhibitor parvulustat (z-2685) from2 streptomyces parvulus
Phyre2
| 15 |
|
PDB 1g94 chain A domain 1
Region: 132 - 145
Aligned: 14
Modelled: 14
Confidence: 6.1%
Identity: 29%
Fold: Glycosyl hydrolase domain
Superfamily: Glycosyl hydrolase domain
Family: alpha-Amylases, C-terminal beta-sheet domain
Phyre2