| 1 |
|
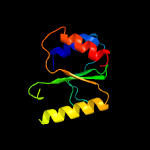
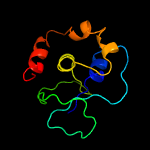
PDB 3r3p chain B
Region: 192 - 291
Aligned: 93
Modelled: 100
Confidence: 99.7%
Identity: 19%
PDB header:hydrolase
Chain: B: PDB Molecule:mobile intron protein;
PDBTitle: homing endonuclease i-bth0305i catalytic domain
Phyre2
| 2 |
|
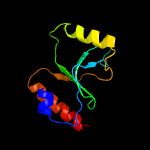
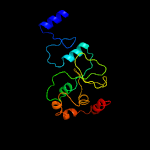
PDB 3hrl chain A
Region: 191 - 292
Aligned: 89
Modelled: 102
Confidence: 99.4%
Identity: 25%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:endonuclease-like protein;
PDBTitle: crystal structure of a putative endonuclease-like protein (ngo0050)2 from neisseria gonorrhoeae
Phyre2
| 3 |
|
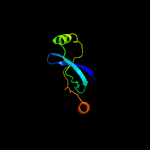
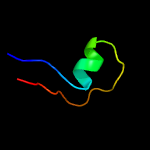
PDB 1cw0 chain A
Region: 190 - 291
Aligned: 96
Modelled: 102
Confidence: 99.3%
Identity: 23%
Fold: Restriction endonuclease-like
Superfamily: Restriction endonuclease-like
Family: Very short patch repair (VSR) endonuclease
Phyre2
| 4 |
|
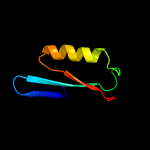
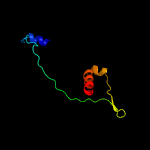
PDB 1vsr chain A
Region: 195 - 293
Aligned: 93
Modelled: 99
Confidence: 98.1%
Identity: 24%
Fold: Restriction endonuclease-like
Superfamily: Restriction endonuclease-like
Family: Very short patch repair (VSR) endonuclease
Phyre2
| 5 |
|
PDB 4oq2 chain A
Region: 197 - 292
Aligned: 95
Modelled: 96
Confidence: 96.2%
Identity: 16%
PDB header:hydrolase
Chain: A: PDB Molecule:restriction endonuclease pvurts1 i;
PDBTitle: 5hmc specific restriction endonuclease pvurts1i
Phyre2
| 6 |
|
PDB 4par chain C
Region: 210 - 291
Aligned: 81
Modelled: 82
Confidence: 96.2%
Identity: 21%
PDB header:dna binding protein/dna
Chain: C: PDB Molecule:uncharacterized protein abasi;
PDBTitle: the 5-hydroxymethylcytosine-specific restriction enzyme abasi in a2 complex with product-like dna
Phyre2
| 7 |
|
PDB 6rdu chain 2
Region: 227 - 276
Aligned: 50
Modelled: 50
Confidence: 95.3%
Identity: 14%
PDB header:proton transport
Chain: 2: PDB Molecule:asa-2: polytomella f-atp synthase associated subunit 2;
PDBTitle: cryo-em structure of polytomella f-atp synthase, rotary substate 1e,2 monomer-masked refinement
Phyre2
| 8 |
|
PDB 1zel chain A domain 2
Region: 71 - 172
Aligned: 100
Modelled: 102
Confidence: 90.7%
Identity: 19%
Fold: Rv2827c C-terminal domain-like
Superfamily: Rv2827c C-terminal domain-like
Family: Rv2827c C-terminal domain-like
Phyre2
| 9 |
|
PDB 1zel chain A
Region: 27 - 172
Aligned: 143
Modelled: 146
Confidence: 89.5%
Identity: 18%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein rv2827c;
PDBTitle: crystal structure of rv2827c protein from mycobacterium tuberculosis
Phyre2
| 10 |
|
PDB 2fcl chain A domain 1
Region: 71 - 96
Aligned: 26
Modelled: 26
Confidence: 54.1%
Identity: 27%
Fold: Nucleotidyltransferase
Superfamily: Nucleotidyltransferase
Family: TM1012-like
Phyre2
| 11 |
|
PDB 1m0d chain A
Region: 193 - 264
Aligned: 64
Modelled: 72
Confidence: 32.9%
Identity: 16%
Fold: Restriction endonuclease-like
Superfamily: Restriction endonuclease-like
Family: Endonuclease I (Holliday junction resolvase)
Phyre2
| 12 |
|
PDB 4gqz chain B
Region: 225 - 243
Aligned: 19
Modelled: 19
Confidence: 15.5%
Identity: 16%
PDB header:metal binding protein
Chain: B: PDB Molecule:putative periplasmic or exported protein;
PDBTitle: crystal structure of s.cuep
Phyre2
| 13 |
|
PDB 2m0n chain A
Region: 282 - 294
Aligned: 13
Modelled: 13
Confidence: 12.2%
Identity: 46%
PDB header:unknown function
Chain: A: PDB Molecule:putative uncharacterized protein;
PDBTitle: solution structure of a duf3349 annotated protein from mycobacterium2 abscessus, mab_3403c. seattle structural genomics center for3 infectious disease target myaba.17112.a.a2
Phyre2
| 14 |
|
PDB 5vrv chain A
Region: 228 - 288
Aligned: 60
Modelled: 61
Confidence: 11.6%
Identity: 17%
PDB header:hydrolase,oxidoreductase
Chain: A: PDB Molecule:protein regulated by acid ph;
PDBTitle: 2.05 angstrom resolution crystal structure of c-terminal domain2 (duf2156) of putative lysylphosphatidylglycerol synthetase from3 agrobacterium fabrum.
Phyre2
| 15 |
|
PDB 2lky chain A
Region: 282 - 294
Aligned: 13
Modelled: 13
Confidence: 9.0%
Identity: 46%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: solution structure of msmeg_1053, the second duf3349 annotated protein2 in the genome of mycobacterium smegmatis, seattle structural genomics3 center for infectious disease target mysma.17112.b
Phyre2
| 16 |
|
PDB 1jg5 chain A
Region: 260 - 273
Aligned: 14
Modelled: 14
Confidence: 7.1%
Identity: 29%
Fold: GTP cyclohydrolase I feedback regulatory protein, GFRP
Superfamily: GTP cyclohydrolase I feedback regulatory protein, GFRP
Family: GTP cyclohydrolase I feedback regulatory protein, GFRP
Phyre2
| 17 |
|
PDB 1tfe chain A
Region: 235 - 291
Aligned: 57
Modelled: 57
Confidence: 6.3%
Identity: 14%
Fold: EF-Ts domain-like
Superfamily: Elongation factor Ts (EF-Ts), dimerisation domain
Family: Elongation factor Ts (EF-Ts), dimerisation domain
Phyre2
| 18 |
|
PDB 4p6i chain B
Region: 33 - 128
Aligned: 74
Modelled: 76
Confidence: 6.0%
Identity: 18%
PDB header:hydrolase
Chain: B: PDB Molecule:crispr-associated endoribonuclease cas2;
PDBTitle: crystal structure of the cas1-cas2 complex from escherichia coli
Phyre2
| 19 |
|
PDB 2vld chain A
Region: 196 - 254
Aligned: 54
Modelled: 56
Confidence: 5.5%
Identity: 19%
PDB header:hydrolase
Chain: A: PDB Molecule:endonuclease nucs;
PDBTitle: crystal structure of a repair endonuclease from pyrococcus abyssi
Phyre2
| 20 |
|
PDB 2csb chain A domain 2
Region: 128 - 146
Aligned: 19
Modelled: 19
Confidence: 5.3%
Identity: 42%
Fold: SAM domain-like
Superfamily: RuvA domain 2-like
Family: Topoisomerase V repeat domain
Phyre2
| 21 |
|
| 22 |
|