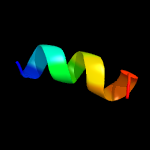
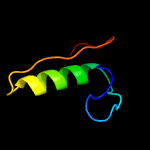
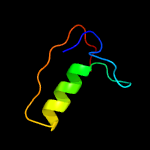
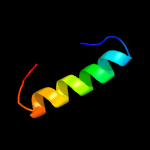
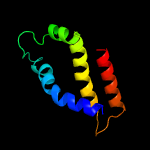
1 c2ctoA_
59.8
50
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: novel protein;PDBTitle: solution structure of the hmg box like domain from human2 hypothetical protein flj14904
2 c4nfuB_
59.4
41
PDB header: signaling proteinChain: B: PDB Molecule: senescence-associated carboxylesterase 101;PDBTitle: structure of the central plant immunity signaling node eds1 in complex2 with its interaction partner sag101
3 c4nfuA_
54.6
38
PDB header: signaling proteinChain: A: PDB Molecule: eds1;PDBTitle: structure of the central plant immunity signaling node eds1 in complex2 with its interaction partner sag101
4 c3ngmB_
35.3
33
PDB header: hydrolaseChain: B: PDB Molecule: extracellular lipase;PDBTitle: crystal structure of lipase from gibberella zeae
5 c3uueA_
35.1
45
PDB header: hydrolaseChain: A: PDB Molecule: lip1, secretory lipase (family 3);PDBTitle: crystal structure of mono- and diacylglycerol lipase from malassezia2 globosa
6 c3o0dF_
33.4
37
PDB header: hydrolaseChain: F: PDB Molecule: triacylglycerol lipase;PDBTitle: crystal structure of lip2 lipase from yarrowia lipolytica at 1.7 a2 resolution
7 c5xk2A_
31.7
32
PDB header: hydrolaseChain: A: PDB Molecule: diacylglycerol lipase;PDBTitle: crystal structure of mono- and diacylglycerol lipase from aspergillus2 oryzae
8 d1tiaa_
27.9
26
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Fungal lipases
9 d1lgya_
27.7
37
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Fungal lipases
10 d3tgla_
25.7
32
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Fungal lipases
11 c6qppA_
25.5
32
PDB header: hydrolaseChain: A: PDB Molecule: lipase;PDBTitle: rhizomucor miehei lipase propeptide complex, native
12 d1tiba_
25.0
53
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Fungal lipases
13 c2kncB_
24.7
26
PDB header: cell adhesionChain: B: PDB Molecule: integrin beta-3;PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
14 c2rmzA_
24.3
26
PDB header: cell adhesionChain: A: PDB Molecule: integrin beta-3;PDBTitle: bicelle-embedded integrin beta3 transmembrane segment
15 c3g7nA_
24.3
32
PDB header: hydrolaseChain: A: PDB Molecule: lipase;PDBTitle: crystal structure of a triacylglycerol lipase from penicillium2 expansum at 1.3
16 c2yijA_
23.6
27
PDB header: hydrolaseChain: A: PDB Molecule: phospholipase a1-iigamma;PDBTitle: crystal structure of phospholipase a1
17 d1kf6d_
23.2
23
Fold: Heme-binding four-helical bundleSuperfamily: Fumarate reductase respiratory complex transmembrane subunitsFamily: Succinate dehydrogenase/Fumarate reductase transmembrane subunits (SdhC/FrdC and SdhD/FrdD)
18 d1dwma_
23.0
60
Fold: CI-2 family of serine protease inhibitorsSuperfamily: CI-2 family of serine protease inhibitorsFamily: CI-2 family of serine protease inhibitors
19 c2ci2I_
22.9
50
PDB header: proteinase inhibitor (chymotrypsin)Chain: I: PDB Molecule: chymotrypsin inhibitor 2;PDBTitle: crystal and molecular structure of the serine proteinase inhibitor ci-2 2 from barley seeds
20 c3rdyA_
22.9
60
PDB header: hydrolase inhibitorChain: A: PDB Molecule: bwi-1=protease inhibitor/trypsin inhibitor;PDBTitle: crystal structure of buckwheat trypsin inhibitor rbti at 1.84 angstrom2 resolution
21 d1to2i_
not modelled
22.9
50
Fold: CI-2 family of serine protease inhibitorsSuperfamily: CI-2 family of serine protease inhibitorsFamily: CI-2 family of serine protease inhibitors
22 d2snii_
not modelled
22.7
50
Fold: CI-2 family of serine protease inhibitorsSuperfamily: CI-2 family of serine protease inhibitorsFamily: CI-2 family of serine protease inhibitors
23 d1ypci_
not modelled
22.6
50
Fold: CI-2 family of serine protease inhibitorsSuperfamily: CI-2 family of serine protease inhibitorsFamily: CI-2 family of serine protease inhibitors
24 c1vbwA_
not modelled
22.6
60
PDB header: protein bindingChain: A: PDB Molecule: trypsin inhibitor bgit;PDBTitle: crystal structure of bitter gourd trypsin inhibitor
25 d1csei_
not modelled
22.5
40
Fold: CI-2 family of serine protease inhibitorsSuperfamily: CI-2 family of serine protease inhibitorsFamily: CI-2 family of serine protease inhibitors
26 c1hymA_
not modelled
22.4
56
PDB header: hydrolase (serine proteinase)Chain: A: PDB Molecule: hydrolyzed cucurbita maxima trypsin inhibitor v;PDBTitle: hydrolyzed trypsin inhibitor (cmti-v, minimized average nmr structure)
27 d1uwca_
not modelled
22.3
47
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Fungal lipases
28 c5gw8A_
not modelled
21.7
32
PDB header: hydrolaseChain: A: PDB Molecule: hypothetical secretory lipase (family 3);PDBTitle: crystal structure of a putative dag-like lipase (mgmdl2) from2 malassezia globosa
29 d1egla_
not modelled
19.6
40
Fold: CI-2 family of serine protease inhibitorsSuperfamily: CI-2 family of serine protease inhibitorsFamily: CI-2 family of serine protease inhibitors
30 c1tinA_
not modelled
18.6
50
PDB header: serine protease inhibitorChain: A: PDB Molecule: trypsin inhibitor v;PDBTitle: three-dimensional structure in solution of cucurbita maxima trypsin2 inhibitor-v determined by nmr spectroscopy
31 c6mhqE_
not modelled
18.2
36
PDB header: membrane proteinChain: E: PDB Molecule: gap junction alpha-3 protein, connexin-46;PDBTitle: structure of connexin-46 intercellular gap junction channel at 3.42 angstrom resolution by cryoem
32 c1njpT_
not modelled
18.0
23
PDB header: ribosomeChain: T: PDB Molecule: general stress protein ctc;PDBTitle: the crystal structure of the 50s large ribosomal subunit2 from deinococcus radiodurans complexed with a trna3 acceptor stem mimic (asm)
33 c3j39M_
not modelled
16.6
38
PDB header: ribosomeChain: M: PDB Molecule: 60s ribosomal protein l14;PDBTitle: structure of the d. melanogaster 60s ribosomal proteins
34 c1cqtI_
not modelled
15.8
35
PDB header: gene regulation/dnaChain: I: PDB Molecule: pou domain, class 2, associating factor 1;PDBTitle: crystal structure of a ternary complex containing an oca-b2 peptide, the oct-1 pou domain, and an octamer element
35 c3jcuj_
not modelled
14.5
25
PDB header: membrane proteinChain: J: PDB Molecule: photosystem ii reaction center protein j;PDBTitle: cryo-em structure of spinach psii-lhcii supercomplex at 3.2 angstrom2 resolution
36 d1rr7a_
not modelled
14.1
28
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Middle operon regulator, Mor
37 c1rr7A_
not modelled
14.1
28
PDB header: transcriptionChain: A: PDB Molecule: middle operon regulator;PDBTitle: crystal structure of the middle operon regulator protein of2 bacteriophage mu
38 c1u1iC_
not modelled
13.7
26
PDB header: isomeraseChain: C: PDB Molecule: myo-inositol-1-phosphate synthase;PDBTitle: myo-inositol phosphate synthase mips from a. fulgidus
39 d1u1ia1
not modelled
13.7
33
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain
40 d2axtj1
not modelled
13.0
20
Fold: Single transmembrane helixSuperfamily: Photosystem II reaction center protein J, PsbJFamily: PsbJ-like
41 c3a0hJ_
not modelled
13.0
20
PDB header: electron transportChain: J: PDB Molecule: photosystem ii reaction center protein j;PDBTitle: crystal structure of i-substituted photosystem ii complex
42 c1cqtJ_
not modelled
12.7
35
PDB header: gene regulation/dnaChain: J: PDB Molecule: pou domain, class 2, associating factor 1;PDBTitle: crystal structure of a ternary complex containing an oca-b2 peptide, the oct-1 pou domain, and an octamer element
43 d3lria_
not modelled
12.6
36
Fold: Insulin-likeSuperfamily: Insulin-likeFamily: Insulin-like
44 c5o9zE_
not modelled
12.4
71
PDB header: splicingChain: E: PDB Molecule: u4/u6 small nuclear ribonucleoprotein prp3;PDBTitle: cryo-em structure of a pre-catalytic human spliceosome primed for2 activation (b complex)
45 c2oryA_
not modelled
12.4
32
PDB header: hydrolaseChain: A: PDB Molecule: lipase;PDBTitle: crystal structure of m37 lipase
46 c5sxpF_
not modelled
11.7
67
PDB header: signaling protein/ligaseChain: F: PDB Molecule: e3 ubiquitin-protein ligase itchy homolog;PDBTitle: structural basis for the interaction between itch prr and beta-pix
47 c1cirA_
not modelled
11.7
63
PDB header: serine protease inhibitorChain: A: PDB Molecule: chymotrypsin inhibitor 2;PDBTitle: complex of two fragments of ci2 [(1-40)(dot)(41-64)]
48 c5sxpG_
not modelled
11.2
67
PDB header: signaling protein/ligaseChain: G: PDB Molecule: e3 ubiquitin-protein ligase itchy homolog;PDBTitle: structural basis for the interaction between itch prr and beta-pix
49 c5ovmA_
not modelled
10.9
32
PDB header: chaperoneChain: A: PDB Molecule: lipase chaperone;PDBTitle: solution structure of lipase binding domain lid1 of foldase from2 pseudomonas aeruginosa
50 c5xnmj_
not modelled
10.8
25
PDB header: membrane proteinChain: J: PDB Molecule: photosystem ii reaction center protein j;PDBTitle: structure of unstacked c2s2m2-type psii-lhcii supercomplex from pisum2 sativum
51 c2mmbA_
not modelled
10.6
42
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: nmr structure of the protein yp_001712342.1 from acinetobacter2 baumannii
52 c6n52B_
not modelled
10.5
13
PDB header: membrane proteinChain: B: PDB Molecule: metabotropic glutamate receptor 5;PDBTitle: metabotropic glutamate receptor 5 apo form
53 c2q5tA_
not modelled
10.4
33
PDB header: toxinChain: A: PDB Molecule: cholix toxin;PDBTitle: full-length cholix toxin from vibrio cholerae
54 c5zazA_
not modelled
10.2
26
PDB header: cell adhesionChain: A: PDB Molecule: integrin beta-2;PDBTitle: solution structure of integrin b2 monomer tranmembrane domain in2 bicelle
55 c1lj2D_
not modelled
9.7
27
PDB header: viral protein/ translationChain: D: PDB Molecule: eukaryotic protein synthesis initiation factor;PDBTitle: recognition of eif4g by rotavirus nsp3 reveals a basis for2 mrna circularization
56 c6igzJ_
not modelled
9.4
33
PDB header: plant proteinChain: J: PDB Molecule: psaj;PDBTitle: structure of psi-lhci
57 c5ys3B_
not modelled
8.9
24
PDB header: transport proteinChain: B: PDB Molecule: succinate-acetate permease;PDBTitle: 1.8 angstrom crystal structure of succinate-acetate permease from2 citrobacter koseri
58 c2lx0A_
not modelled
8.9
55
PDB header: membrane proteinChain: A: PDB Molecule: membrane fusion protein p14;PDBTitle: arced helix (arch) nmr structure of the reovirus p14 fusion-associated2 small transmembrane (fast) protein transmembrane domain (tmd) in3 dodecyl phosphocholine (dpc) micelles
59 c2qjkM_
not modelled
8.9
12
PDB header: electron transportChain: M: PDB Molecule: cytochrome b;PDBTitle: crystal structure analysis of mutant rhodobacter2 sphaeroides bc1 with stigmatellin and antimycin
60 d2qam01
not modelled
8.4
20
Fold: Rubredoxin-likeSuperfamily: Zn-binding ribosomal proteinsFamily: Ribosomal protein L32p
61 c2mwqA_
not modelled
8.3
57
PDB header: plant proteinChain: A: PDB Molecule: oxygen-evolving enhancer protein 3, chloroplastic;PDBTitle: solution structure of psbq from spinacia oleracea
62 c6mx7C_
not modelled
8.2
45
PDB header: virusChain: C: PDB Molecule: capsid;PDBTitle: cryoem structure of chimeric eastern equine encephalitis virus:2 genome-binding capsid n-terminal domain
63 c6epcN_
not modelled
7.7
15
PDB header: hydrolaseChain: N: PDB Molecule: 26s proteasome non-atpase regulatory subunit 1;PDBTitle: ground state 26s proteasome (gs2)
64 c5fg0B_
not modelled
7.5
14
PDB header: ligaseChain: B: PDB Molecule: e3 ubiquitin-protein ligase listerin;PDBTitle: structure of the conserved yeast listerin (ltn1) n-terminal domain,2 monoclinic form
65 c3j3v0_
not modelled
7.5
36
PDB header: ribosomeChain: 0: PDB Molecule: 50s ribosomal protein l32;PDBTitle: atomic model of the immature 50s subunit from bacillus subtilis (state2 i-a)
66 c6hu9U_
not modelled
7.4
19
PDB header: oxidoreductase/electron transportChain: U: PDB Molecule: cytochrome b-c1 complex subunit 10;PDBTitle: iii2-iv2 mitochondrial respiratory supercomplex from s. cerevisiae
67 c5mlc7_
not modelled
7.3
47
PDB header: ribosomeChain: 7: PDB Molecule: psrp5alpha, chloroplastic;PDBTitle: cryo-em structure of the spinach chloroplast ribosome reveals the2 location of plastid-specific ribosomal proteins and extensions
68 d1dc1a_
not modelled
7.2
32
Fold: Restriction endonuclease-likeSuperfamily: Restriction endonuclease-likeFamily: Restriction endonuclease BsobI
69 c5xfsA_
not modelled
7.1
26
PDB header: protein transportChain: A: PDB Molecule: pe family protein pe8;PDBTitle: crystal structure of pe8-ppe15 in complex with espg5 from m.2 tuberculosis
70 c1y4eA_
not modelled
7.1
57
PDB header: membrane proteinChain: A: PDB Molecule: sodium/hydrogen exchanger 1;PDBTitle: nmr structure of transmembrane segment iv of the nhe12 isoform of the na+/h+ exchanger
71 c6o4mB_
not modelled
7.0
16
PDB header: toxinChain: B: PDB Molecule: melittin;PDBTitle: racemic melittin
72 c2mltB_
not modelled
7.0
16
PDB header: toxin (hemolytic polypeptide)Chain: B: PDB Molecule: melittin;PDBTitle: melittin
73 c2mw6A_
not modelled
7.0
16
PDB header: toxinChain: A: PDB Molecule: melittin;PDBTitle: structure of the bee venom toxin melittin with [(c5h5)ru]+ fragment2 attached to the tryptophan residue
74 c6o4mA_
not modelled
7.0
16
PDB header: toxinChain: A: PDB Molecule: melittin;PDBTitle: racemic melittin
75 c6dstA_
not modelled
7.0
16
PDB header: toxinChain: A: PDB Molecule: melittin;PDBTitle: recombinant melittin
76 c2mltA_
not modelled
7.0
16
PDB header: toxin (hemolytic polypeptide)Chain: A: PDB Molecule: melittin;PDBTitle: melittin
77 c2g38A_
not modelled
6.9
11
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: pe family protein;PDBTitle: a pe/ppe protein complex from mycobacterium tuberculosis
78 d2g38a1
not modelled
6.9
11
Fold: Ferritin-likeSuperfamily: PE/PPE dimer-likeFamily: PE
79 c1pjmA_
not modelled
6.8
100
PDB header: protein transportChain: A: PDB Molecule: retinoblastoma-associated protein;PDBTitle: mouse importin alpha-bipartite nls from human2 retinoblastoma protein complex
80 c4b1yM_
not modelled
6.7
46
PDB header: structural proteinChain: M: PDB Molecule: phosphatase and actin regulator 1;PDBTitle: structure of the phactr1 rpel-3 bound to g-actin
81 c2mckA_
not modelled
6.7
44
PDB header: hydrolaseChain: A: PDB Molecule: polyprotein;PDBTitle: backbone 1h, 13c, and 15n chemical shift assignments for murine2 norovirus cr6 ns1/2 protein
82 c5h1sg_
not modelled
6.6
54
PDB header: ribosomeChain: G: PDB Molecule: 50s ribosomal protein l4, chloroplastic;PDBTitle: structure of the large subunit of the chloro-ribosome
83 d2j0151
not modelled
6.6
50
Fold: Rubredoxin-likeSuperfamily: Zn-binding ribosomal proteinsFamily: Ribosomal protein L32p
84 c5zghJ_
not modelled
6.5
25
PDB header: photosynthesisChain: J: PDB Molecule: psaj;PDBTitle: cryo-em structure of the red algal psi-lhcr
85 d2e74h1
not modelled
6.4
54
Fold: Single transmembrane helixSuperfamily: PetN subunit of the cytochrome b6f complexFamily: PetN subunit of the cytochrome b6f complex
86 c5vnyA_
not modelled
6.4
63
PDB header: endocytosis, protein bindingChain: A: PDB Molecule: lethal (2) giant discs 1, isoform b;PDBTitle: crystal structure of dm14-3 domain of lgd
87 c6e3yP_
not modelled
6.3
38
PDB header: signaling proteinChain: P: PDB Molecule: calcitonin gene-related peptide 1;PDBTitle: cryo-em structure of the active, gs-protein complexed, human cgrp2 receptor
88 c4i7zH_
not modelled
6.3
54
PDB header: photosynthesisChain: H: PDB Molecule: cytochrome b6-f complex subunit 8;PDBTitle: crystal structure of cytochrome b6f in dopg, with disordered rieske2 iron-sulfur protein soluble domain
89 c4pv1H_
not modelled
6.3
54
PDB header: electron transport/inhibitorChain: H: PDB Molecule: cytochrome b6-f complex subunit 8;PDBTitle: cytochrome b6f structure from m. laminosus with the quinone analog2 inhibitor stigmatellin
90 c2kncA_
not modelled
6.3
30
PDB header: cell adhesionChain: A: PDB Molecule: integrin alpha-iib;PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
91 c5wu5C_
not modelled
6.3
27
PDB header: transferaseChain: C: PDB Molecule: speckle targeted pip5k1a-regulated poly(a) polymerase;PDBTitle: crystal structure of apo human tut1, form iii
92 c3pq1A_
not modelled
6.3
27
PDB header: transferaseChain: A: PDB Molecule: poly(a) rna polymerase;PDBTitle: crystal structure of human mitochondrial poly(a) polymerase (papd1)
93 c6gwxA_
not modelled
6.2
75
PDB header: structural proteinChain: A: PDB Molecule: optimised ppa-tyr;PDBTitle: stabilising and understanding a miniprotein by rational design.
94 c5f5uH_
not modelled
6.2
38
PDB header: splicingChain: H: PDB Molecule: putative uncharacterized protein;PDBTitle: crystal structure of the snu23-prp38-mfap1(217-258) complex of2 chaetomium thermophilum
95 c1z8yI_
not modelled
6.2
35
PDB header: virusChain: I: PDB Molecule: spike glycoprotein e1;PDBTitle: mapping the e2 glycoprotein of alphaviruses
96 c1z8yM_
not modelled
6.2
35
PDB header: virusChain: M: PDB Molecule: spike glycoprotein e1;PDBTitle: mapping the e2 glycoprotein of alphaviruses
97 c1z8yK_
not modelled
6.2
35
PDB header: virusChain: K: PDB Molecule: spike glycoprotein e1;PDBTitle: mapping the e2 glycoprotein of alphaviruses
98 c1z8yO_
not modelled
6.2
35
PDB header: virusChain: O: PDB Molecule: spike glycoprotein e1;PDBTitle: mapping the e2 glycoprotein of alphaviruses
99 c2e75H_
not modelled
6.2
54
PDB header: photosynthesisChain: H: PDB Molecule: cytochrome b6-f complex subunit 8;PDBTitle: crystal structure of the cytochrome b6f complex with 2-nonyl-4-2 hydroxyquinoline n-oxide (nqno) from m.laminosus