| 1 |
|
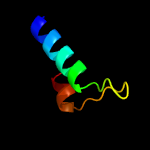
PDB 5xfs chain B
Region: 2 - 81
Aligned: 80
Modelled: 80
Confidence: 100.0%
Identity: 39%
PDB header:protein transport
Chain: B: PDB Molecule:ppe family protein ppe15;
PDBTitle: crystal structure of pe8-ppe15 in complex with espg5 from m.2 tuberculosis
Phyre2
| 2 |
|
PDB 2g38 chain B domain 1
Region: 2 - 82
Aligned: 81
Modelled: 81
Confidence: 100.0%
Identity: 23%
Fold: Ferritin-like
Superfamily: PE/PPE dimer-like
Family: PPE
Phyre2
| 3 |
|
PDB 2g38 chain B
Region: 2 - 82
Aligned: 81
Modelled: 81
Confidence: 100.0%
Identity: 23%
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:ppe family protein;
PDBTitle: a pe/ppe protein complex from mycobacterium tuberculosis
Phyre2
| 4 |
|
PDB 4xy3 chain A
Region: 3 - 79
Aligned: 77
Modelled: 77
Confidence: 99.9%
Identity: 13%
PDB header:protein transport
Chain: A: PDB Molecule:esx-1 secretion-associated protein espb;
PDBTitle: structure of esx-1 secreted protein espb
Phyre2
| 5 |
|
PDB 4xb6 chain D
Region: 19 - 53
Aligned: 17
Modelled: 17
Confidence: 43.4%
Identity: 35%
PDB header:transferase
Chain: D: PDB Molecule:alpha-d-ribose 1-methylphosphonate 5-phosphate c-p lyase;
PDBTitle: structure of the e. coli c-p lyase core complex
Phyre2
| 6 |
|
PDB 6aok chain A
Region: 44 - 51
Aligned: 8
Modelled: 7
Confidence: 21.0%
Identity: 38%
PDB header:hydrolase
Chain: A: PDB Molecule:ceg4;
PDBTitle: crystal structure of legionella pneumophila effector ceg4 with n-2 terminal tev protease cleavage sequence
Phyre2
| 7 |
|
PDB 1zee chain A domain 1
Region: 30 - 53
Aligned: 24
Modelled: 23
Confidence: 20.8%
Identity: 38%
Fold: Indolic compounds 2,3-dioxygenase-like
Superfamily: Indolic compounds 2,3-dioxygenase-like
Family: Indoleamine 2,3-dioxygenase-like
Phyre2
| 8 |
|
PDB 6cgj chain A
Region: 44 - 51
Aligned: 8
Modelled: 8
Confidence: 19.8%
Identity: 38%
PDB header:hydrolase
Chain: A: PDB Molecule:effector protein lem4 (lpg1101);
PDBTitle: structure of the had domain of effector protein lem4 (lpg1101) from2 legionella pneumophila
Phyre2
| 9 |
|
PDB 1bzg chain A
Region: 50 - 56
Aligned: 7
Modelled: 7
Confidence: 14.5%
Identity: 14%
PDB header:hormone
Chain: A: PDB Molecule:parathyroid hormone-related protein;
PDBTitle: the solution structure of human parathyroid hormone-related2 protein (1-34) in near-physiological solution, nmr, 303 structures
Phyre2
| 10 |
|
PDB 2nvj chain A
Region: 44 - 53
Aligned: 10
Modelled: 10
Confidence: 12.8%
Identity: 40%
PDB header:hydrolase
Chain: A: PDB Molecule:25mer peptide from vacuolar atp synthase subunit
PDBTitle: nmr structures of transmembrane segment from subunit a from2 the yeast proton v-atpase
Phyre2
| 11 |
|
PDB 3sjr chain B
Region: 21 - 35
Aligned: 15
Modelled: 9
Confidence: 11.5%
Identity: 20%
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of conserved unkown function protein cv_1783 from2 chromobacterium violaceum atcc 12472
Phyre2
| 12 |
|
PDB 1vlf chain N domain 1
Region: 31 - 35
Aligned: 5
Modelled: 5
Confidence: 10.7%
Identity: 100%
Fold: Prealbumin-like
Superfamily: Cna protein B-type domain
Family: Cna protein B-type domain
Phyre2
| 13 |
|
PDB 5l85 chain B
Region: 28 - 38
Aligned: 11
Modelled: 11
Confidence: 10.2%
Identity: 36%
PDB header:signaling protein
Chain: B: PDB Molecule:nuclear fragile x mental retardation-interacting protein 1;
PDBTitle: solution structure of the complex between human znhit3 and nufip12 proteins
Phyre2
| 14 |
|
PDB 6nbi chain P
Region: 52 - 56
Aligned: 5
Modelled: 5
Confidence: 9.5%
Identity: 80%
PDB header:signaling protein
Chain: P: PDB Molecule:long-acting parathyroid hormone analog;
PDBTitle: cryo-em structure of parathyroid hormone receptor type 1 in complex2 with a long-acting parathyroid hormone analog and g protein
Phyre2
| 15 |
|
PDB 2jtw chain A
Region: 46 - 53
Aligned: 8
Modelled: 8
Confidence: 8.4%
Identity: 50%
PDB header:membrane protein
Chain: A: PDB Molecule:transmembrane helix 7 of yeast vatpase;
PDBTitle: solution structure of tm7 bound to dpc micelles
Phyre2
| 16 |
|
PDB 1fcd chain A domain 3
Region: 45 - 58
Aligned: 14
Modelled: 14
Confidence: 8.1%
Identity: 29%
Fold: CO dehydrogenase flavoprotein C-domain-like
Superfamily: FAD/NAD-linked reductases, dimerisation (C-terminal) domain
Family: FAD/NAD-linked reductases, dimerisation (C-terminal) domain
Phyre2
| 17 |
|
PDB 6qpi chain A
Region: 22 - 54
Aligned: 32
Modelled: 33
Confidence: 7.0%
Identity: 25%
PDB header:membrane protein
Chain: A: PDB Molecule:anoctamin-6;
PDBTitle: cryo-em structure of calcium-free mtmem16f lipid scramblase in2 nanodisc
Phyre2
| 18 |
|
PDB 4c47 chain B
Region: 297 - 305
Aligned: 9
Modelled: 9
Confidence: 6.0%
Identity: 22%
PDB header:cell adhesion
Chain: B: PDB Molecule:inner membrane lipoprotein;
PDBTitle: salmonella enterica trimeric lipoprotein sadb
Phyre2