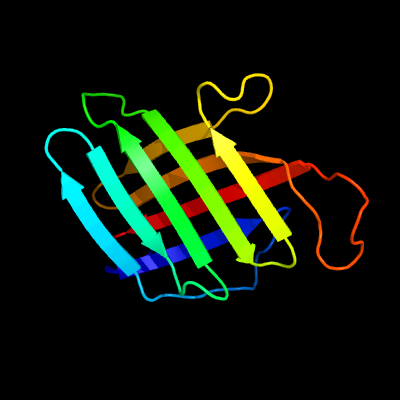
1 c4xinB_
100.0
34
PDB header: unknown functionChain: B: PDB Molecule: lpqh orthologue;PDBTitle: x-ray crystal structure of an lpqh orthologue from mycobacterium avium
2 c4zjmA_
100.0
90
PDB header: unknown functionChain: A: PDB Molecule: lipoprotein lpqh;PDBTitle: crystal structure of mycobacterium tuberculosis lpqh (rv3763)
3 c6qt9Y_
61.1
13
PDB header: virusChain: Y: PDB Molecule: orf 31;PDBTitle: cryo-em structure of sh1 full particle.
4 c2g16A_
28.6
15
PDB header: luminescent proteinChain: A: PDB Molecule: green fluorescent protein;PDBTitle: structure of s65a y66s gfp variant after backbone2 fragmentation
5 d1wv3a1
25.5
44
Fold: SMAD/FHA domainSuperfamily: SMAD/FHA domainFamily: EssC N-terminal domain-like
6 c2gw4C_
21.2
18
PDB header: luminescent proteinChain: C: PDB Molecule: kaede;PDBTitle: crystal structure of stony coral fluorescent protein kaede, red form
7 d2icha1
20.1
19
Fold: AttH-likeSuperfamily: AttH-likeFamily: AttH-like
8 c6fx6A_
19.5
11
PDB header: unknown functionChain: A: PDB Molecule: satie-ted;PDBTitle: thioester domain of the staphylococcus aureus tie protein
9 c6h9cb_
17.7
18
PDB header: virusChain: B: PDB Molecule: vp4;PDBTitle: cryo-em structure of archaeal extremophilic internal membrane-2 containing haloarcula californiae icosahedral virus 1 (hciv-1) at3 3.74 angstroms resolution.
10 c3najA_
17.3
19
PDB header: sugar binding proteinChain: A: PDB Molecule: galectin-8;PDBTitle: crystal structure of a protease-resistant mutant form of human2 galectin-8
11 c3ff7B_
16.5
18
PDB header: cell adhesion/immune systemChain: B: PDB Molecule: epithelial cadherin;PDBTitle: structure of nk cell receptor klrg1 bound to e-cadherin
12 c1vraA_
15.6
19
PDB header: transferaseChain: A: PDB Molecule: arginine biosynthesis bifunctional protein argj;PDBTitle: crystal structure of arginine biosynthesis bifunctional protein argj2 (10175521) from bacillus halodurans at 2.00 a resolution
13 c2mraA_
15.2
20
PDB header: de novo proteinChain: A: PDB Molecule: de novo designed protein or459;PDBTitle: solution nmr structure of de novo designed protein, northeast2 structural genomics consortium (nesg) target or459
14 c3wp6A_
14.4
24
PDB header: hydrolaseChain: A: PDB Molecule: cdbfv;PDBTitle: the complex structure of cdbfv e109a with xylotriose
15 c3lf4A_
13.7
18
PDB header: fluorescent proteinChain: A: PDB Molecule: fluorescent timer precursor blue102;PDBTitle: crystal structure of fluorescent timer precursor blue102
16 c3mxnB_
13.4
14
PDB header: replicationChain: B: PDB Molecule: recq-mediated genome instability protein 2;PDBTitle: crystal structure of the rmi core complex
17 c3c12A_
12.8
17
PDB header: biosynthetic proteinChain: A: PDB Molecule: flagellar protein;PDBTitle: crystal structure of flgd from xanthomonas campestris:2 insights into the hook capping essential for flagellar3 assembly
18 c2ovsB_
12.7
4
PDB header: gene regulation, ligand binding proteinChain: B: PDB Molecule: l0044;PDBTitle: crystal strcuture of a type three secretion system protein
19 c4m02A_
12.6
22
PDB header: calcium binding proteinChain: A: PDB Molecule: serine-rich adhesin for platelets;PDBTitle: middle fragment(residues 494-663) of the binding region of srap
20 d1mywa_
11.4
11
Fold: GFP-likeSuperfamily: GFP-likeFamily: Fluorescent proteins
21 c3rwaE_
not modelled
9.9
21
PDB header: fluorescent proteinChain: E: PDB Molecule: fluorescent protein fp480;PDBTitle: crystal structure of circular-permutated mkate
22 d2go8a1
not modelled
9.7
32
Fold: Ferredoxin-likeSuperfamily: Dimeric alpha+beta barrelFamily: PG130-like
23 d1sgoa_
not modelled
9.5
29
Fold: N domain of copper amine oxidase-likeSuperfamily: Hypothetical protein c14orf129, hspc210Family: Hypothetical protein c14orf129, hspc210
24 d1kp5a_
not modelled
9.4
15
Fold: GFP-likeSuperfamily: GFP-likeFamily: Fluorescent proteins
25 c2l7yA_
not modelled
9.1
20
PDB header: structural proteinChain: A: PDB Molecule: putative endo-beta-n-acetylglucosaminidase;PDBTitle: solution structure of a putative surface protein
26 c2a56A_
not modelled
9.0
38
PDB header: luminescent proteinChain: A: PDB Molecule: gfp-like non-fluorescent chromoprotein fp595 chain 1;PDBTitle: fluorescent protein asfp595, a143s, on-state, 5min irradiation
27 c2jtyA_
not modelled
8.5
24
PDB header: structural proteinChain: A: PDB Molecule: type-1 fimbrial protein, a chain;PDBTitle: self-complemented variant of fima, the main subunit of type 1 pilus
28 c5az3A_
not modelled
8.1
21
PDB header: transport proteinChain: A: PDB Molecule: abc-type transporter, periplasmic component;PDBTitle: crystal structure of heme binding protein hmut
29 c2q9kA_
not modelled
8.1
14
PDB header: oxidoreductaseChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a putative oxidoreductase (exig_1997) from2 exiguobacterium sibiricum 255-15 at 1.59 a resolution
30 c3akoG_
not modelled
8.0
11
PDB header: fluorescent proteinChain: G: PDB Molecule: venus;PDBTitle: crystal structure of the reassembled venus
31 c4p7hB_
not modelled
8.0
21
PDB header: motor/fluorescent proteinChain: B: PDB Molecule: myosin-7,green fluorescent protein;PDBTitle: structure of human beta-cardiac myosin motor domain::gfp chimera
32 c4gpvB_
not modelled
7.9
14
PDB header: cell adhesionChain: B: PDB Molecule: putative cell adhesion protein;PDBTitle: crystal structure of a putative cell adhesion protein (bacegg_00536)2 from bacteroides eggerthii dsm 20697 at 1.67 a resolution
33 c2mblA_
not modelled
7.9
39
PDB header: de novo proteinChain: A: PDB Molecule: top7 fold protein top7m13;PDBTitle: solution nmr structure of de novo designed top7 fold protein top7m13,2 northeast structural genomics consortium (nesg) target or33
34 c4uznA_
not modelled
7.7
35
PDB header: hydrolaseChain: A: PDB Molecule: endo-beta-1,4-glucanase (celulase b);PDBTitle: the native structure of the family 46 carbohydrate-binding2 module (cbm46) of endo-beta-1,4-glucanase b (cel5b) from3 bacillus halodurans
35 c3u0kA_
not modelled
7.5
25
PDB header: fluorescent proteinChain: A: PDB Molecule: rcamp;PDBTitle: crystal structure of the genetically encoded calcium indicator rcamp
36 d1rx0a2
not modelled
7.1
50
Fold: Acyl-CoA dehydrogenase NM domain-likeSuperfamily: Acyl-CoA dehydrogenase NM domain-likeFamily: Medium chain acyl-CoA dehydrogenase, NM (N-terminal and middle) domains
37 c5u75A_
not modelled
7.1
29
PDB header: toxinChain: A: PDB Molecule: enterotoxin-like toxin x;PDBTitle: the structure of staphylococcal enterotoxin-like x (selx), a unique2 superantigen
38 c3t9gB_
not modelled
7.0
24
PDB header: lyaseChain: B: PDB Molecule: pectate lyase;PDBTitle: the crystal structure of family 3 pectate lyase from2 caldicellulosiruptor bescii
39 c4i2yB_
not modelled
6.9
18
PDB header: fluorescent proteinChain: B: PDB Molecule: rgeco1;PDBTitle: crystal structure of the genetically encoded calcium indicator rgeco1
40 c2m5sA_
not modelled
6.6
19
PDB header: viral proteinChain: A: PDB Molecule: coat protein;PDBTitle: high-resolution nmr structure and cryo-em imaging support multiple2 functional roles for the accessory i-domain of phage p22 coat protein
41 c2bgpA_
not modelled
6.5
12
PDB header: carbohydrate binding proteinChain: A: PDB Molecule: endo-b1,4-mannanase 5c;PDBTitle: mannan binding module from man5c in bound conformation
42 d2d29a2
not modelled
6.5
26
Fold: Acyl-CoA dehydrogenase NM domain-likeSuperfamily: Acyl-CoA dehydrogenase NM domain-likeFamily: Medium chain acyl-CoA dehydrogenase, NM (N-terminal and middle) domains
43 c4anjA_
not modelled
6.4
15
PDB header: motor protein/metal-bindng proteinChain: A: PDB Molecule: unconventional myosin-vi, green fluorescent protein;PDBTitle: myosin vi (mdinsert2-gfp fusion) pre-powerstroke state (mg.adp.alf4)
44 c3evpA_
not modelled
6.3
14
PDB header: signaling proteinChain: A: PDB Molecule: green fluorescent protein,green fluorescent protein;PDBTitle: crystal structure of circular-permutated egfp
45 c3k25B_
not modelled
6.0
19
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: slr1438 protein;PDBTitle: crystal structure of slr1438 protein from synechocystis sp. pcc 6803,2 northeast structural genomics consortium target sgr112
46 c4l3aA_
not modelled
5.9
18
PDB header: cell invasionChain: A: PDB Molecule: internalin k;PDBTitle: crystal structure of internalin k (inlk) from listeria monocytogenes
47 c3uv0B_
not modelled
5.7
17
PDB header: protein bindingChain: B: PDB Molecule: mutator 2, isoform b;PDBTitle: crystal structure of the drosophila mu2 fha domain
48 c3uv1B_
not modelled
5.6
13
PDB header: allergenChain: B: PDB Molecule: der f 7 allergen;PDBTitle: crystal structure a major allergen from dust mite
49 c6efrA_
not modelled
5.3
18
PDB header: choline-binding proteinChain: A: PDB Molecule: inicsnfr 1.0, a genetically encoded nicotine biosensor,PDBTitle: crystal structure of inicsnfr 1.0
50 c3h3iA_
not modelled
5.3
56
PDB header: lipid binding proteinChain: A: PDB Molecule: putative lipid binding protein;PDBTitle: crystal structure of a putative lipid binding protein (bt_2261) from2 bacteroides thetaiotaomicron vpi-5482 at 2.20 a resolution
51 c4ktoB_
not modelled
5.2
21
PDB header: oxidoreductaseChain: B: PDB Molecule: isovaleryl-coa dehydrogenase;PDBTitle: crystal structure of a putative isovaleryl-coa dehydrogenase (psi-2 nysgrc-012251) from sinorhizobium meliloti 1021
52 c6dgvA_
not modelled
5.1
18
PDB header: fluorescent proteinChain: A: PDB Molecule: fluorescent gaba sensor precursor;PDBTitle: igabasnfr fluorescent gaba sensor precursor
53 c4ndkA_
not modelled
5.1
14
PDB header: fluorescent protein, de novo proteinChain: A: PDB Molecule: e23p-yfp, gfp-like fluorescent chromoprotein fp506,PDBTitle: crystal structure of a computational designed engrailed homeodomain2 variant fused with yfp