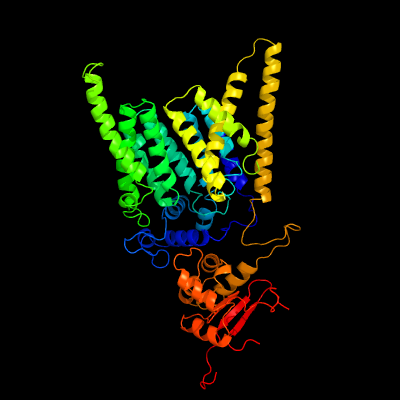
| 1 | c6eznF_
|
|
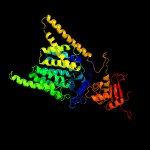 |
99.9 |
14 |
PDB header:membrane protein
Chain: F: PDB Molecule:dolichyl-diphosphooligosaccharide--protein
PDBTitle: cryo-em structure of the yeast oligosaccharyltransferase (ost) complex
|
|
|
|
| 2 | c3wajA_
|
|
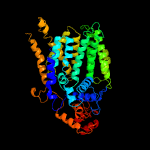 |
99.8 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:transmembrane oligosaccharyl transferase;
PDBTitle: crystal structure of the archaeoglobus fulgidus2 oligosaccharyltransferase (o29867_arcfu) complex with zn and sulfate
|
|
|
|
| 3 | c3rceA_
|
|
 |
99.6 |
12 |
PDB header:transferase/peptide
Chain: A: PDB Molecule:oligosaccharide transferase to n-glycosylate proteins;
PDBTitle: bacterial oligosaccharyltransferase pglb
|
|
|
|
| 4 | c5f15A_
|
|
 |
99.4 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:4-amino-4-deoxy-l-arabinose (l-ara4n) transferase;
PDBTitle: crystal structure of arnt from cupriavidus metallidurans bound to2 undecaprenyl phosphate
|
|
|
|
| 5 | c3vu0B_
|
|
 |
98.5 |
15 |
PDB header:transferase
Chain: B: PDB Molecule:putative uncharacterized protein;
PDBTitle: crystal structure of the c-terminal globular domain of2 oligosaccharyltransferase (afaglb-s2, af_0040, o30195_arcfu) from3 archaeoglobus fulgidus
|
|
|
|
| 6 | c3vgpA_
|
|
 |
98.3 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:transmembrane oligosaccharyl transferase, putative;
PDBTitle: crystal structure of the c-terminal globular domain of2 oligosaccharyltransferase (af_0329) from archaeoglobus fulgidus
|
|
|
|
| 7 | c6p2rB_
|
|
 |
96.3 |
13 |
PDB header:transferase
Chain: B: PDB Molecule:dolichyl-phosphate-mannose--protein mannosyltransferase 2;
PDBTitle: structure of s. cerevisiae protein o-mannosyltransferase pmt1-pmt22 complex bound to the sugar donor
|
|
|
|
| 8 | c6p25A_
|
|
 |
95.5 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:dolichyl-phosphate-mannose--protein mannosyltransferase 1;
PDBTitle: structure of s. cerevisiae protein o-mannosyltransferase pmt1-pmt22 complex bound to the sugar donor and a peptide acceptor
|
|
|
|
| 9 | c2lgzA_
|
|
 |
94.2 |
11 |
PDB header:transferase, membrane protein
Chain: A: PDB Molecule:dolichyl-diphosphooligosaccharide--protein
PDBTitle: solution structure of stt3p
|
|
|
|
| 10 | c3waiA_
|
|
 |
92.6 |
16 |
PDB header:transferase, transport protein
Chain: A: PDB Molecule:maltose-binding periplasmic protein, transmembrane
PDBTitle: crystal structure of the c-terminal globular domain of2 oligosaccharyltransferase (afaglb-l, o29867_arcfu) from archaeoglobus3 fulgidus as a mbp fusion
|
|
|
|
| 11 | c3vu1A_
|
|
 |
83.0 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:putative uncharacterized protein ph0242;
PDBTitle: crystal structure of the c-terminal globular domain of2 oligosaccharyltransferase (phaglb-l, o74088_pyrho) from pyrococcus3 horikoshii
|
|
|
|
| 12 | c2nacA_
|
|
 |
67.5 |
13 |
PDB header:oxidoreductase(aldehyde(d),nad+(a))
Chain: A: PDB Molecule:nad-dependent formate dehydrogenase;
PDBTitle: high resolution structures of holo and apo formate dehydrogenase
|
|
|
|
| 13 | c5svbD_
|
|
 |
57.2 |
16 |
PDB header:ligase
Chain: D: PDB Molecule:acetone carboxylase alpha subunit;
PDBTitle: mechanism of atp-dependent acetone carboxylation, acetone carboxylase2 amp bound structure
|
|
|
|
| 14 | c5l9wA_
|
|
 |
55.8 |
10 |
PDB header:ligase
Chain: A: PDB Molecule:acetophenone carboxylase delta subunit;
PDBTitle: crystal structure of the apc core complex
|
|
|
|
| 15 | c3fn4A_
|
|
 |
52.4 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nad-dependent formate dehydrogenase;
PDBTitle: apo-form of nad-dependent formate dehydrogenase from bacterium2 moraxella sp.c-1 in closed conformation
|
|
|
|
| 16 | c2b5dX_
|
|
 |
50.6 |
4 |
PDB header:hydrolase
Chain: X: PDB Molecule:alpha-amylase;
PDBTitle: crystal structure of the novel alpha-amylase amyc from thermotoga2 maritima
|
|
|
|
| 17 | d2naca2
|
|
 |
46.5 |
13 |
Fold:Flavodoxin-like
Superfamily:Formate/glycerate dehydrogenase catalytic domain-like
Family:Formate/glycerate dehydrogenases, substrate-binding domain |
|
|
|
| 18 | d2odgc1
|
|
 |
40.6 |
19 |
Fold:LEM/SAP HeH motif
Superfamily:LEM domain
Family:LEM domain |
|
|
|
| 19 | c2hyxA_
|
|
 |
35.8 |
15 |
PDB header:unknown function
Chain: A: PDB Molecule:protein dipz;
PDBTitle: structure of the c-terminal domain of dipz from mycobacterium2 tuberculosis
|
|
|
|
| 20 | d1c01a_
|
|
 |
34.8 |
21 |
Fold:gamma-Crystallin-like
Superfamily:gamma-Crystallin-like
Family:Plant antimicrobial protein MIAMP1 |
|
|
|
| 21 | c3fo5A_ |
|
not modelled |
33.5 |
22 |
PDB header:lipid transport
Chain: A: PDB Molecule:thioesterase, adipose associated, isoform bfit2;
PDBTitle: human start domain of acyl-coenzyme a thioesterase 11 (acot11)
|
|
|
| 22 | d1t9ba2 |
|
not modelled |
29.5 |
10 |
Fold:Thiamin diphosphate-binding fold (THDP-binding)
Superfamily:Thiamin diphosphate-binding fold (THDP-binding)
Family:Pyruvate oxidase and decarboxylase Pyr module |
|
|
| 23 | c3qocD_ |
|
not modelled |
27.9 |
21 |
PDB header:hydrolase
Chain: D: PDB Molecule:putative metallopeptidase;
PDBTitle: crystal structure of n-terminal domain (creatinase/prolidase like2 domain) of putative metallopeptidase from corynebacterium diphtheriae
|
|
|
| 24 | d1ufaa2 |
|
not modelled |
27.4 |
18 |
Fold:7-stranded beta/alpha barrel
Superfamily:Glycoside hydrolase/deacetylase
Family:AmyC N-terminal domain-like |
|
|
| 25 | c2xznU_ |
|
not modelled |
25.4 |
20 |
PDB header:ribosome
Chain: U: PDB Molecule:ribosomal protein l7ae containing protein;
PDBTitle: crystal structure of the eukaryotic 40s ribosomal2 subunit in complex with initiation factor 1. this file3 contains the 40s subunit and initiation factor for4 molecule 2
|
|
|
| 26 | c2opkC_ |
|
not modelled |
24.5 |
5 |
PDB header:isomerase
Chain: C: PDB Molecule:hypothetical protein;
PDBTitle: crystal structure of a putative mannose-6-phosphate isomerase2 (reut_a1446) from ralstonia eutropha jmp134 at 2.10 a resolution
|
|
|
| 27 | c4hzbE_ |
|
not modelled |
22.6 |
11 |
PDB header:hydrolase
Chain: E: PDB Molecule:putative periplasmic protein;
PDBTitle: crystal structure of the type vi semet effector-immunity complex tae3-2 tai3 from ralstonia pickettii
|
|
|
| 28 | c5t0zB_ |
|
not modelled |
22.4 |
22 |
PDB header:structural protein
Chain: B: PDB Molecule:lipoprotein, putative;
PDBTitle: pelc from geobacter metallireducens
|
|
|
| 29 | c2zagB_ |
|
not modelled |
21.4 |
14 |
PDB header:transferase
Chain: B: PDB Molecule:oligosaccharyl transferase stt3 subunit related protein;
PDBTitle: crystal structure of the semet-substituted soluble domain of stt3 from2 p. furiosus
|
|
|
| 30 | d1xzwa2 |
|
not modelled |
21.2 |
6 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:Purple acid phosphatase-like |
|
|
| 31 | c2miiA_ |
|
not modelled |
20.8 |
8 |
PDB header:protein binding
Chain: A: PDB Molecule:penicillin-binding protein activator lpob;
PDBTitle: nmr structure of e. coli lpob
|
|
|
| 32 | c3zk4A_ |
|
not modelled |
20.5 |
9 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:diphosphonucleotide phosphatase 1;
PDBTitle: structure of purple acid phosphatase ppd1 isolated from2 yellow lupin (lupinus luteus) seeds
|
|
|
| 33 | d1pvda2 |
|
not modelled |
19.8 |
7 |
Fold:Thiamin diphosphate-binding fold (THDP-binding)
Superfamily:Thiamin diphosphate-binding fold (THDP-binding)
Family:Pyruvate oxidase and decarboxylase Pyr module |
|
|
| 34 | c2ou2A_ |
|
not modelled |
19.6 |
29 |
PDB header:transferase
Chain: A: PDB Molecule:histone acetyltransferase htatip;
PDBTitle: acetyltransferase domain of human hiv-1 tat interacting2 protein, 60kda, isoform 3
|
|
|
| 35 | d2ozua1 |
|
not modelled |
19.2 |
24 |
Fold:Acyl-CoA N-acyltransferases (Nat)
Superfamily:Acyl-CoA N-acyltransferases (Nat)
Family:N-acetyl transferase, NAT |
|
|
| 36 | c5ujwD_ |
|
not modelled |
18.9 |
14 |
PDB header:isomerase
Chain: D: PDB Molecule:triosephosphate isomerase;
PDBTitle: crystal structure of triosephosphate isomerase from francisella2 tularensis subsp. tularensis schu s4
|
|
|
| 37 | c3zey1_ |
|
not modelled |
18.7 |
9 |
PDB header:ribosome
Chain: 1: PDB Molecule:40s ribosomal protein s4, putative;
PDBTitle: high-resolution cryo-electron microscopy structure of the trypanosoma2 brucei ribosome
|
|
|
| 38 | d1fy7a_ |
|
not modelled |
18.6 |
24 |
Fold:Acyl-CoA N-acyltransferases (Nat)
Superfamily:Acyl-CoA N-acyltransferases (Nat)
Family:N-acetyl transferase, NAT |
|
|
| 39 | c5oxwD_ |
|
not modelled |
18.5 |
24 |
PDB header:splicing
Chain: D: PDB Molecule:neq068;
PDBTitle: structure of neqn from nanoarchaeum equitans
|
|
|
| 40 | c5azaA_ |
|
not modelled |
18.5 |
19 |
PDB header:sugar binding protein, transferase
Chain: A: PDB Molecule:maltose-binding periplasmic protein,oligosaccharyl
PDBTitle: crystal structure of mbp-saglb fusion protein with a 20-residue spacer2 in the connector helix
|
|
|
| 41 | d2b5dx2 |
|
not modelled |
18.4 |
4 |
Fold:7-stranded beta/alpha barrel
Superfamily:Glycoside hydrolase/deacetylase
Family:AmyC N-terminal domain-like |
|
|
| 42 | d1zpda2 |
|
not modelled |
18.4 |
14 |
Fold:Thiamin diphosphate-binding fold (THDP-binding)
Superfamily:Thiamin diphosphate-binding fold (THDP-binding)
Family:Pyruvate oxidase and decarboxylase Pyr module |
|
|
| 43 | c2givA_ |
|
not modelled |
18.3 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:probable histone acetyltransferase myst1;
PDBTitle: human myst histone acetyltransferase 1
|
|
|
| 44 | d2giva1 |
|
not modelled |
18.3 |
19 |
Fold:Acyl-CoA N-acyltransferases (Nat)
Superfamily:Acyl-CoA N-acyltransferases (Nat)
Family:N-acetyl transferase, NAT |
|
|
| 45 | c1ufaA_ |
|
not modelled |
18.0 |
18 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:tt1467 protein;
PDBTitle: crystal structure of tt1467 from thermus thermophilus hb8
|
|
|
| 46 | c3izbD_ |
|
not modelled |
17.8 |
12 |
PDB header:ribosome
Chain: D: PDB Molecule:40s ribosomal protein rps4 (s4e);
PDBTitle: localization of the small subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
|
|
|
| 47 | d3bfxa1 |
|
not modelled |
17.8 |
20 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:PAPS sulfotransferase |
|
|
| 48 | c5gk9A_ |
|
not modelled |
17.5 |
29 |
PDB header:transferase/metal binding protein
Chain: A: PDB Molecule:histone acetyltransferase kat7;
PDBTitle: crystal structure of human hbo1 in complex with brpf2
|
|
|
| 49 | d1ovma2 |
|
not modelled |
17.4 |
14 |
Fold:Thiamin diphosphate-binding fold (THDP-binding)
Superfamily:Thiamin diphosphate-binding fold (THDP-binding)
Family:Pyruvate oxidase and decarboxylase Pyr module |
|
|
| 50 | d2pjua1 |
|
not modelled |
16.9 |
9 |
Fold:Chelatase-like
Superfamily:PrpR receptor domain-like
Family:PrpR receptor domain-like |
|
|
| 51 | c4hrvB_ |
|
not modelled |
16.7 |
11 |
PDB header:lipid binding protein
Chain: B: PDB Molecule:putative lipoprotein gna1162;
PDBTitle: crystal structure of lipoprotein gna1162 from neisseria meningitidis
|
|
|
| 52 | c2xwgA_ |
|
not modelled |
16.3 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:sortase;
PDBTitle: crystal structure of sortase c-1 from actinomyces oris (formerly2 actinomyces naeslundii)
|
|
|
| 53 | d2z06a1 |
|
not modelled |
16.2 |
12 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:TTHA0625-like |
|
|
| 54 | c4g1kB_ |
|
not modelled |
15.8 |
9 |
PDB header:isomerase
Chain: B: PDB Molecule:triosephosphate isomerase;
PDBTitle: crystal structure of triosephosphate isomerase from burkholderia2 thailandensis
|
|
|
| 55 | c4x9tA_ |
|
not modelled |
15.6 |
10 |
PDB header:unknown function
Chain: A: PDB Molecule:uncharacterized protein upf0065;
PDBTitle: crystal structure of a tctc solute binding protein from polaromonas2 (bpro_3516, target efi-510338), no ligand
|
|
|
| 56 | d1w2za3 |
|
not modelled |
15.4 |
11 |
Fold:Cystatin-like
Superfamily:Amine oxidase N-terminal region
Family:Amine oxidase N-terminal region |
|
|
| 57 | d2obba1 |
|
not modelled |
15.1 |
9 |
Fold:HAD-like
Superfamily:HAD-like
Family:BT0820-like |
|
|
| 58 | c2q5cA_ |
|
not modelled |
15.0 |
7 |
PDB header:transcription
Chain: A: PDB Molecule:ntrc family transcriptional regulator;
PDBTitle: crystal structure of ntrc family transcriptional regulator from2 clostridium acetobutylicum
|
|
|
| 59 | c3j20E_ |
|
not modelled |
14.8 |
11 |
PDB header:ribosome
Chain: E: PDB Molecule:30s ribosomal protein s4e;
PDBTitle: promiscuous behavior of proteins in archaeal ribosomes revealed by2 cryo-em: implications for evolution of eukaryotic ribosomes (30s3 ribosomal subunit)
|
|
|
| 60 | c3n92A_ |
|
not modelled |
14.6 |
4 |
PDB header:transferase
Chain: A: PDB Molecule:alpha-amylase, gh57 family;
PDBTitle: crystal structure of tk1436, a gh57 branching enzyme from2 hyperthermophilic archaeon thermococcus kodakaraensis, in complex3 with glucose
|
|
|
| 61 | d1r5aa2 |
|
not modelled |
14.4 |
16 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione S-transferase (GST), N-terminal domain |
|
|
| 62 | c5ohxB_ |
|
not modelled |
14.4 |
8 |
PDB header:lyase
Chain: B: PDB Molecule:cystathionine beta-synthase;
PDBTitle: structure of active cystathionine b-synthase from apis mellifera
|
|
|
| 63 | c5essB_ |
|
not modelled |
14.2 |
15 |
PDB header:transferase
Chain: B: PDB Molecule:2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-
PDBTitle: crystal structure of m. tuberculosis mend bound to mg2+ and covalent2 intermediate i (a thdp and decarboxylated 2-oxoglutarate adduct)
|
|
|
| 64 | c4y90B_ |
|
not modelled |
14.0 |
0 |
PDB header:isomerase
Chain: B: PDB Molecule:triosephosphate isomerase;
PDBTitle: crystal structure of triosephosphate isomerase from deinococcus2 radiodurans
|
|
|
| 65 | d1suxa_ |
|
not modelled |
13.9 |
11 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
|
|
| 66 | c2x58B_ |
|
not modelled |
13.9 |
14 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:peroxisomal bifunctional enzyme;
PDBTitle: the crystal structure of mfe1 liganded with coa
|
|
|
| 67 | d1sw3a_ |
|
not modelled |
13.7 |
5 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
|
|
| 68 | c4y8fA_ |
|
not modelled |
13.6 |
11 |
PDB header:isomerase
Chain: A: PDB Molecule:triosephosphate isomerase;
PDBTitle: crystal structure of triosephosphate isomerase from clostridium2 perfringens
|
|
|
| 69 | c4mknA_ |
|
not modelled |
13.6 |
5 |
PDB header:isomerase
Chain: A: PDB Molecule:triosephosphate isomerase;
PDBTitle: crystal structure of chloroplastic triosephosphate isomerase from2 chlamydomonas reinhardtii at 1.1 a of resolution
|
|
|
| 70 | c6bveA_ |
|
not modelled |
13.5 |
11 |
PDB header:isomerase
Chain: A: PDB Molecule:triosephosphate isomerase;
PDBTitle: triosephosphate isomerase of synechocystis in complex with 2-2 phosphoglycolic acid
|
|
|
| 71 | c3th6B_ |
|
not modelled |
13.5 |
5 |
PDB header:isomerase
Chain: B: PDB Molecule:triosephosphate isomerase;
PDBTitle: crystal structure of triosephosphate isomerase from rhipicephalus2 (boophilus) microplus.
|
|
|
| 72 | c2jgqB_ |
|
not modelled |
13.5 |
17 |
PDB header:isomerase
Chain: B: PDB Molecule:triosephosphate isomerase;
PDBTitle: kinetics and structural properties of triosephosphate2 isomerase from helicobacter pylori
|
|
|
| 73 | c5zg5B_ |
|
not modelled |
13.4 |
0 |
PDB header:isomerase
Chain: B: PDB Molecule:triosephosphate isomerase;
PDBTitle: crystal structure of triosephosphate isomerase sadsubaaa mutant from2 opisthorchis viverrini
|
|
|
| 74 | d1kv5a_ |
|
not modelled |
13.3 |
11 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
|
|
| 75 | c5xyiE_ |
|
not modelled |
13.2 |
12 |
PDB header:ribosome
Chain: E: PDB Molecule:40s ribosomal protein s4;
PDBTitle: small subunit of trichomonas vaginalis ribosome
|
|
|
| 76 | c1kbpB_ |
|
not modelled |
13.0 |
7 |
PDB header:hydrolase (phosphoric monoester)
Chain: B: PDB Molecule:purple acid phosphatase;
PDBTitle: kidney bean purple acid phosphatase
|
|
|
| 77 | c2pjuD_ |
|
not modelled |
13.0 |
9 |
PDB header:transcription
Chain: D: PDB Molecule:propionate catabolism operon regulatory protein;
PDBTitle: crystal structure of propionate catabolism operon regulatory protein2 prpr
|
|
|
| 78 | d1o5xa_ |
|
not modelled |
12.9 |
22 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
|
|
| 79 | d1n55a_ |
|
not modelled |
12.9 |
0 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
|
|
| 80 | c4nvtD_ |
|
not modelled |
12.9 |
5 |
PDB header:isomerase
Chain: D: PDB Molecule:triosephosphate isomerase;
PDBTitle: crystal structure of triosephosphate isomerase from brucella2 melitensis
|
|
|
| 81 | c5eywB_ |
|
not modelled |
12.9 |
0 |
PDB header:isomerase
Chain: B: PDB Molecule:triosephosphate isomerase;
PDBTitle: crystal structure of litopenaeus vannamei triosephosphate isomerase2 complexed with 2-phosphoglycolic acid
|
|
|
| 82 | d1r2ra_ |
|
not modelled |
12.6 |
5 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
|
|
| 83 | c5ibxB_ |
|
not modelled |
12.4 |
5 |
PDB header:isomerase
Chain: B: PDB Molecule:triosephosphate isomerase;
PDBTitle: 1.65 angstrom crystal structure of triosephosphate isomerase (tim)2 from streptococcus pneumoniae
|
|
|
| 84 | d1neya_ |
|
not modelled |
12.4 |
11 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
|
|
| 85 | d1kfia4 |
|
not modelled |
12.3 |
18 |
Fold:TBP-like
Superfamily:Phosphoglucomutase, C-terminal domain
Family:Phosphoglucomutase, C-terminal domain |
|
|
| 86 | d2btma_ |
|
not modelled |
12.2 |
11 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
|
|
| 87 | c1yyaA_ |
|
not modelled |
12.2 |
16 |
PDB header:isomerase
Chain: A: PDB Molecule:triosephosphate isomerase;
PDBTitle: crystal structure of tt0473, putative triosephosphate isomerase from2 thermus thermophilus hb8
|
|
|
| 88 | c6faqA_ |
|
not modelled |
12.2 |
13 |
PDB header:dna binding protein
Chain: A: PDB Molecule:dna binding protein;
PDBTitle: structure of h. salinarum rosr (vng0258) grown from kbr
|
|
|
| 89 | c5dynA_ |
|
not modelled |
12.2 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative peptidase;
PDBTitle: b. fragilis cysteine protease
|
|
|
| 90 | c3m9yB_ |
|
not modelled |
12.2 |
16 |
PDB header:isomerase
Chain: B: PDB Molecule:triosephosphate isomerase;
PDBTitle: crystal structure of triosephosphate isomerase from methicillin2 resistant staphylococcus aureus at 1.9 angstrom resolution
|
|
|
| 91 | d3pmga4 |
|
not modelled |
12.2 |
25 |
Fold:TBP-like
Superfamily:Phosphoglucomutase, C-terminal domain
Family:Phosphoglucomutase, C-terminal domain |
|
|
| 92 | c3u5cE_ |
|
not modelled |
12.2 |
12 |
PDB header:ribosome
Chain: E: PDB Molecule:40s ribosomal protein s4-a;
PDBTitle: the structure of the eukaryotic ribosome at 3.0 a resolution. this2 entry contains proteins of the 40s subunit, ribosome a
|
|
|
| 93 | c4y9aB_ |
|
not modelled |
12.2 |
11 |
PDB header:isomerase
Chain: B: PDB Molecule:triosephosphate isomerase;
PDBTitle: crystal structure of triosephosphate isomerase from streptomyces2 coelicolor
|
|
|
| 94 | d1mo0a_ |
|
not modelled |
12.1 |
11 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
|
|
| 95 | d1vpqa_ |
|
not modelled |
12.1 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:TM1631-like
Family:TM1631-like |
|
|
| 96 | c5uprA_ |
|
not modelled |
12.1 |
11 |
PDB header:isomerase
Chain: A: PDB Molecule:triosephosphate isomerase;
PDBTitle: x-ray structure of a putative triosephosphate isomerase from2 toxoplasma gondii me49
|
|
|
| 97 | d1b9ba_ |
|
not modelled |
12.1 |
11 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
|
|
| 98 | d1m6ja_ |
|
not modelled |
12.0 |
5 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
|
|
| 99 | c3s6dA_ |
|
not modelled |
12.0 |
0 |
PDB header:isomerase
Chain: A: PDB Molecule:putative triosephosphate isomerase;
PDBTitle: crystal structure of a putative triosephosphate isomerase from2 coccidioides immitis
|
|
|